Table of Contents
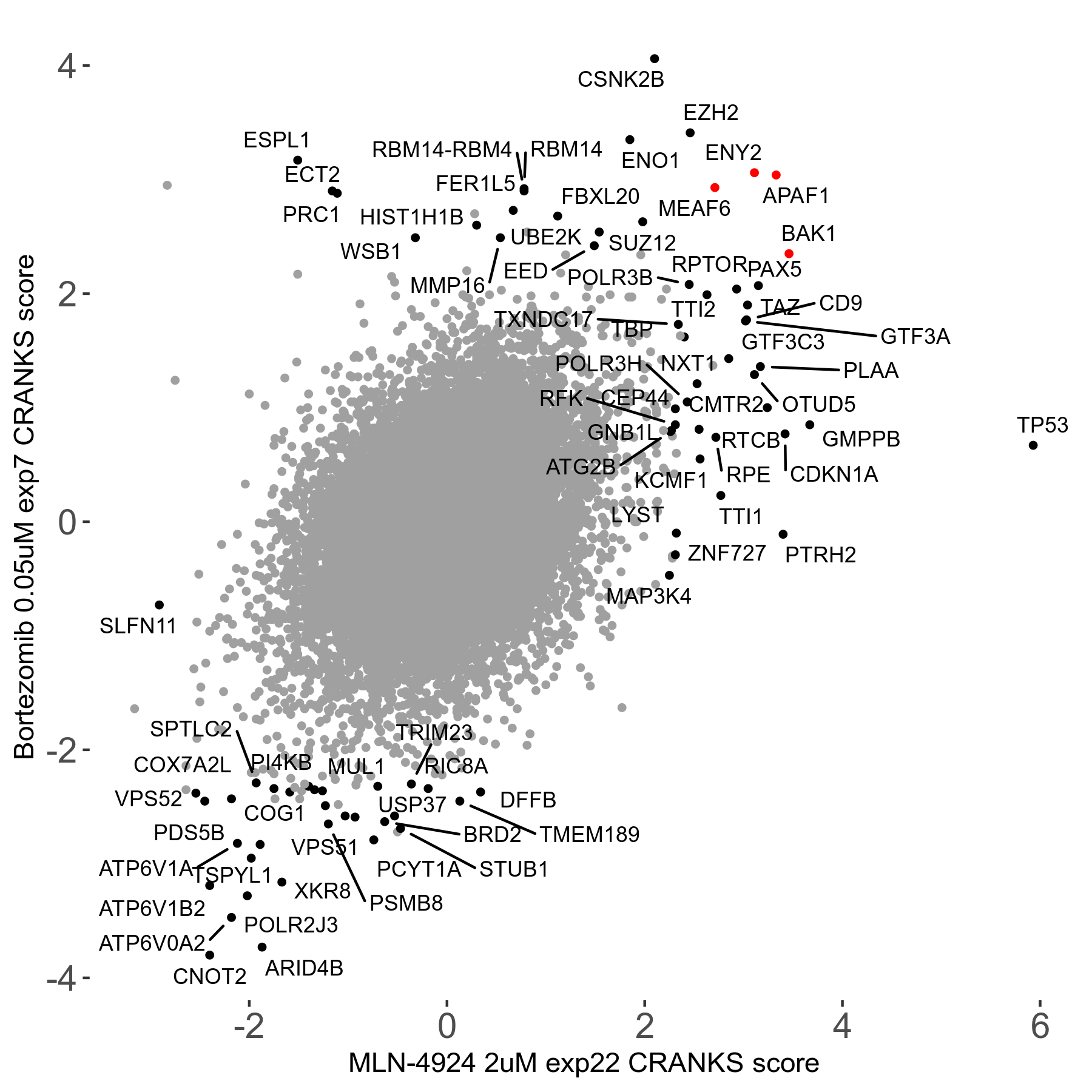
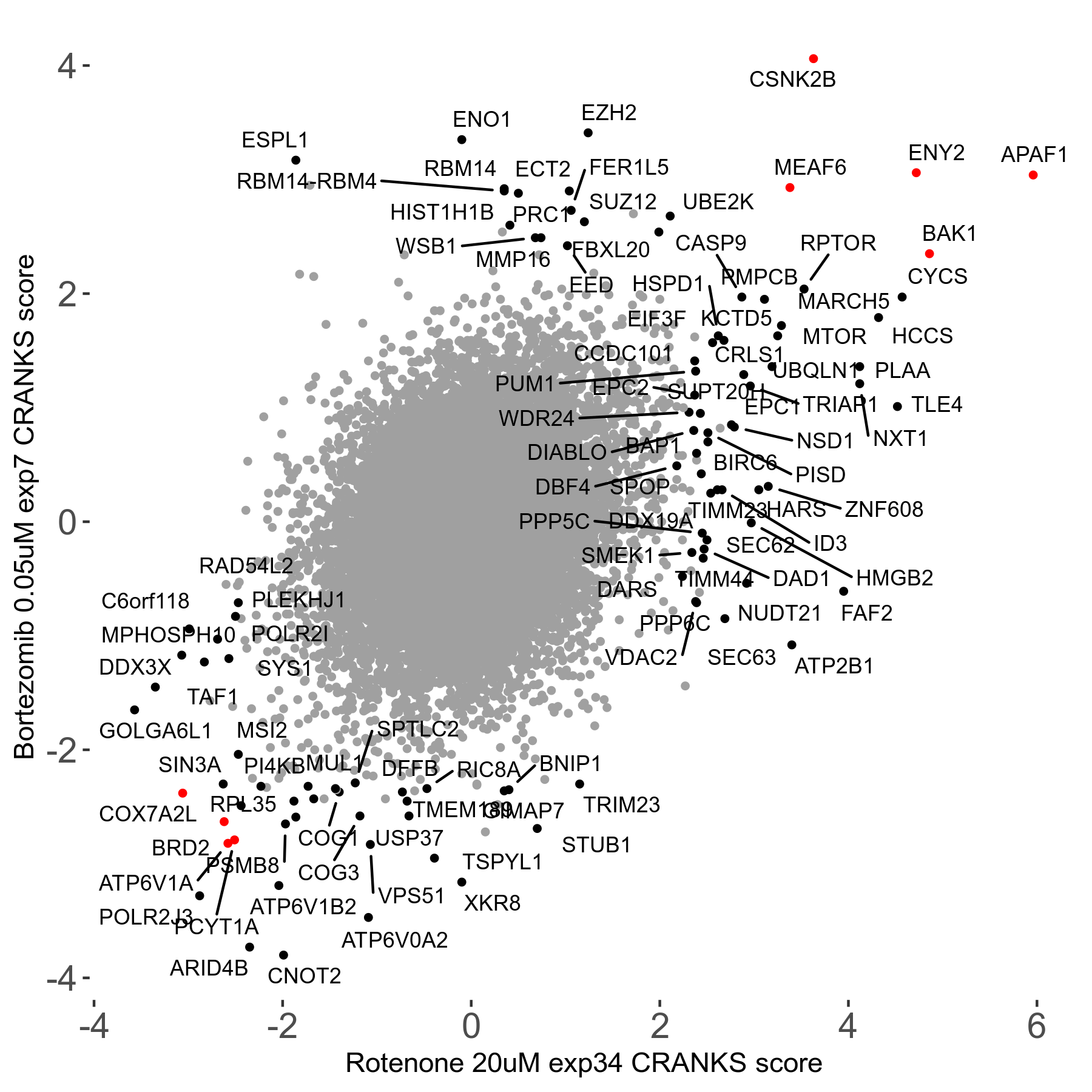
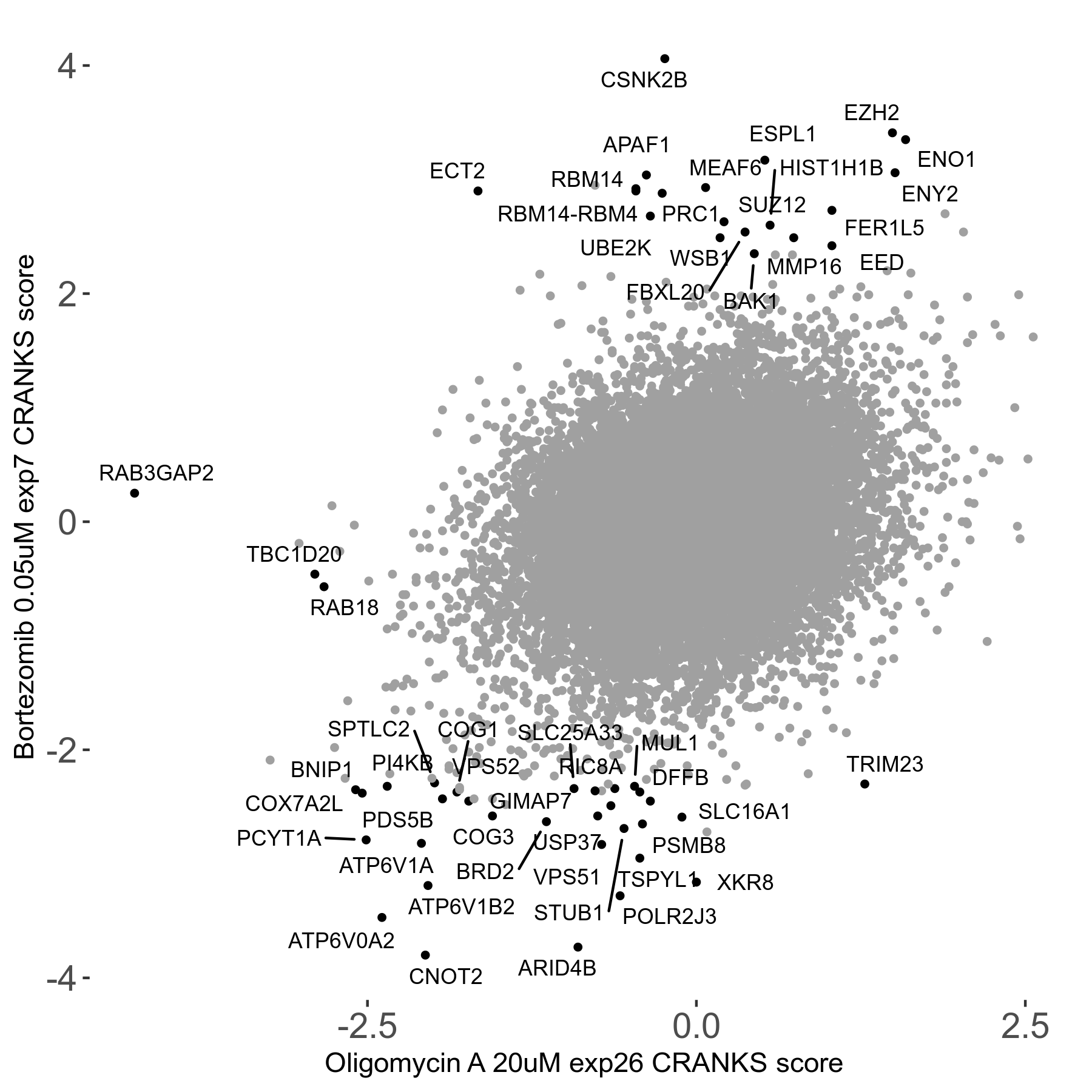
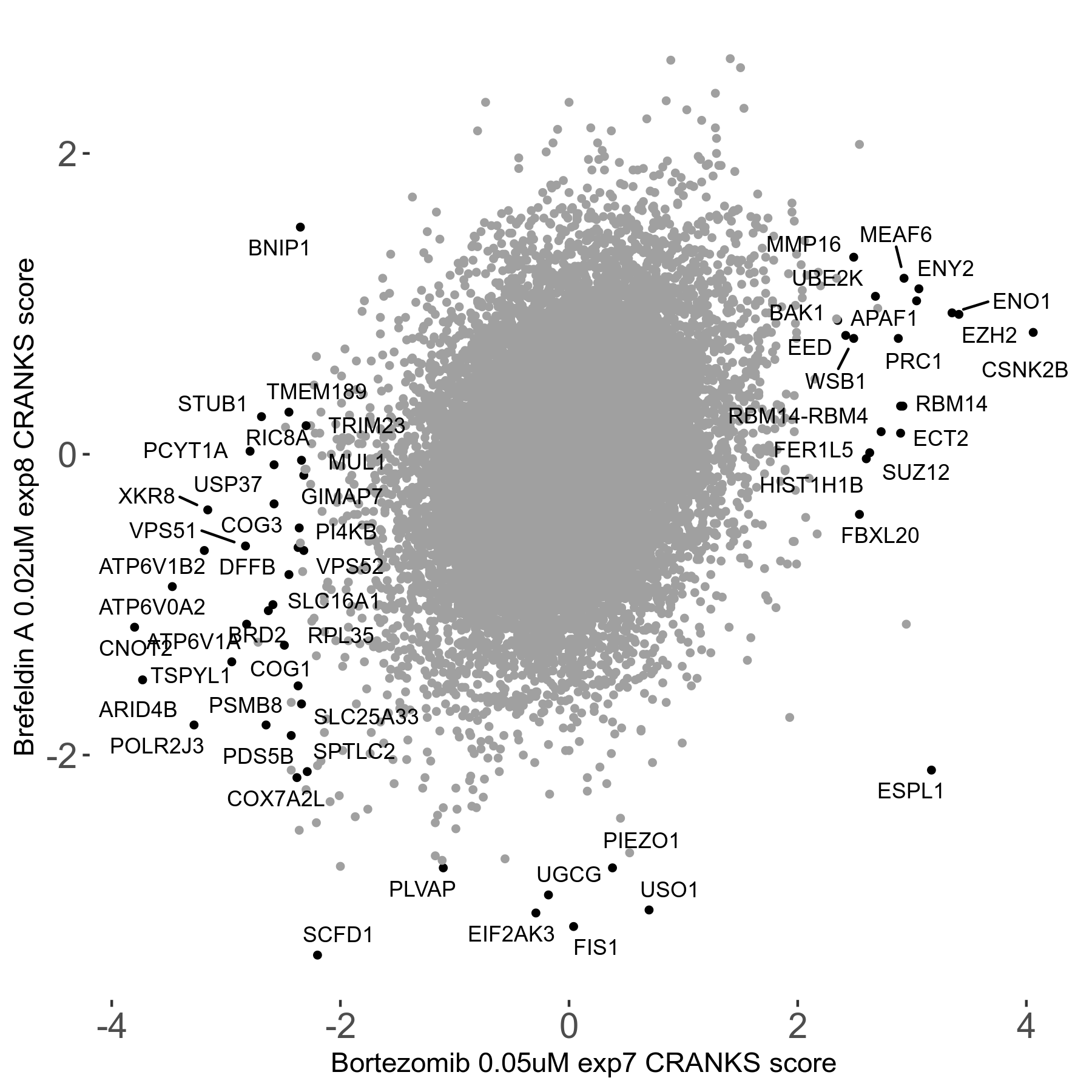
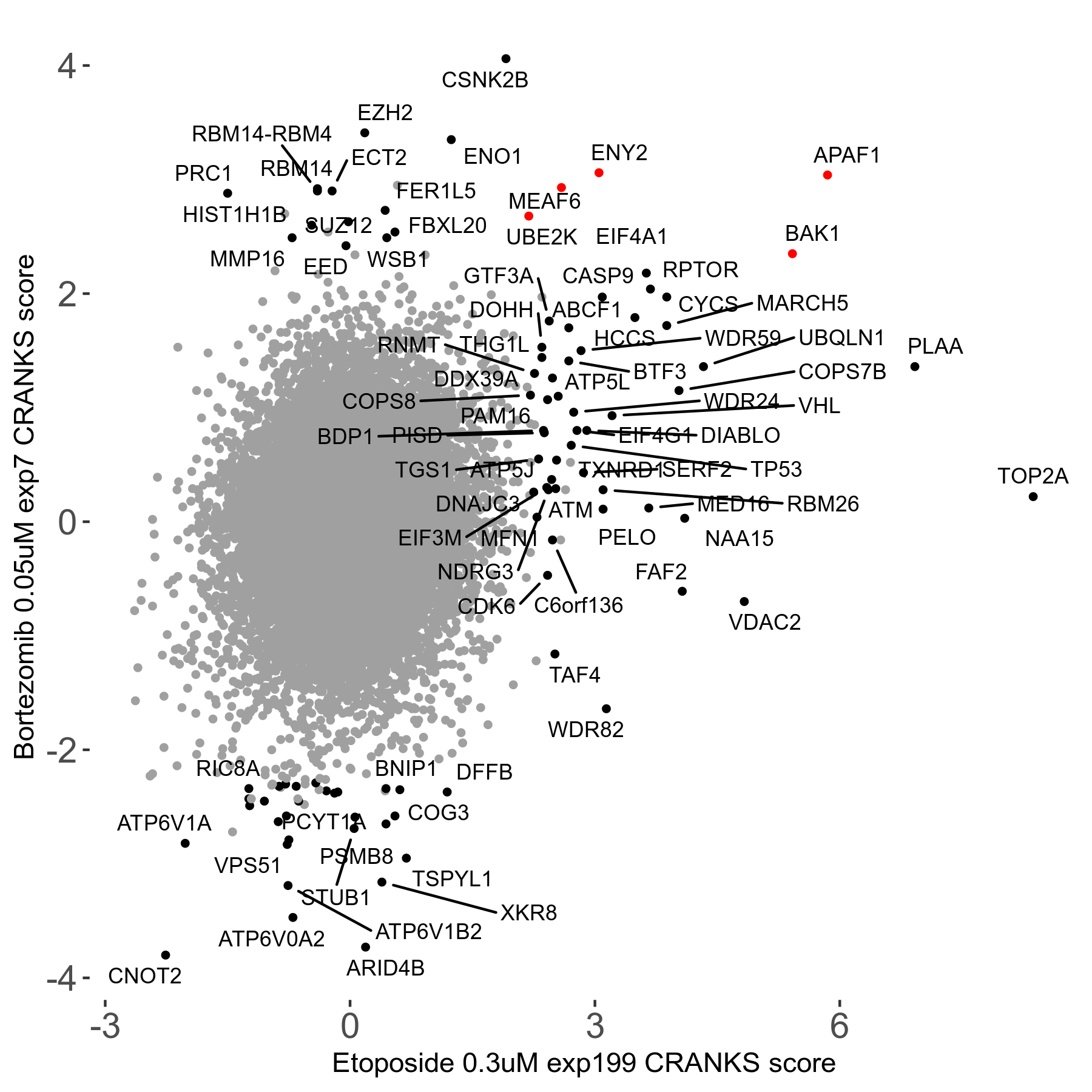
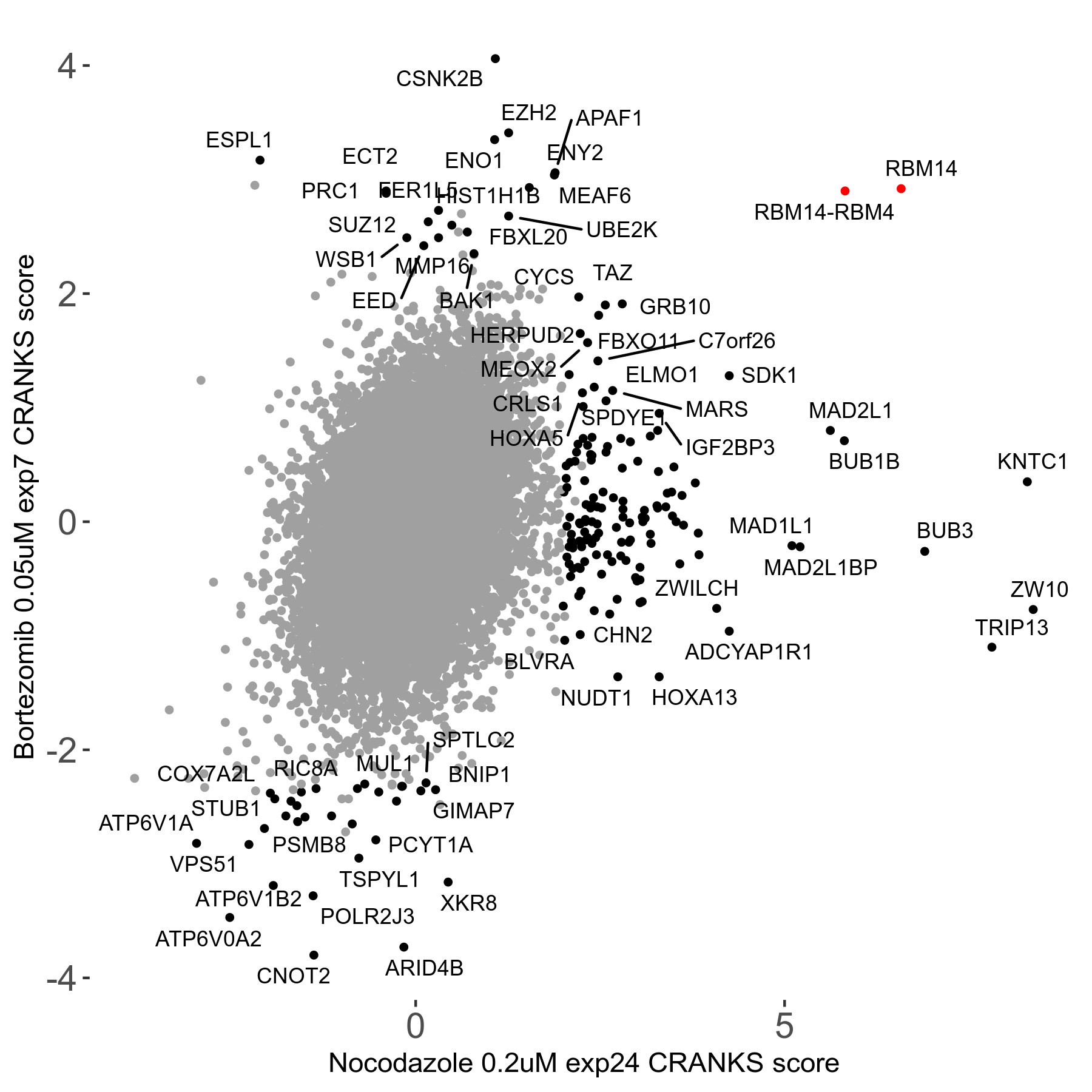
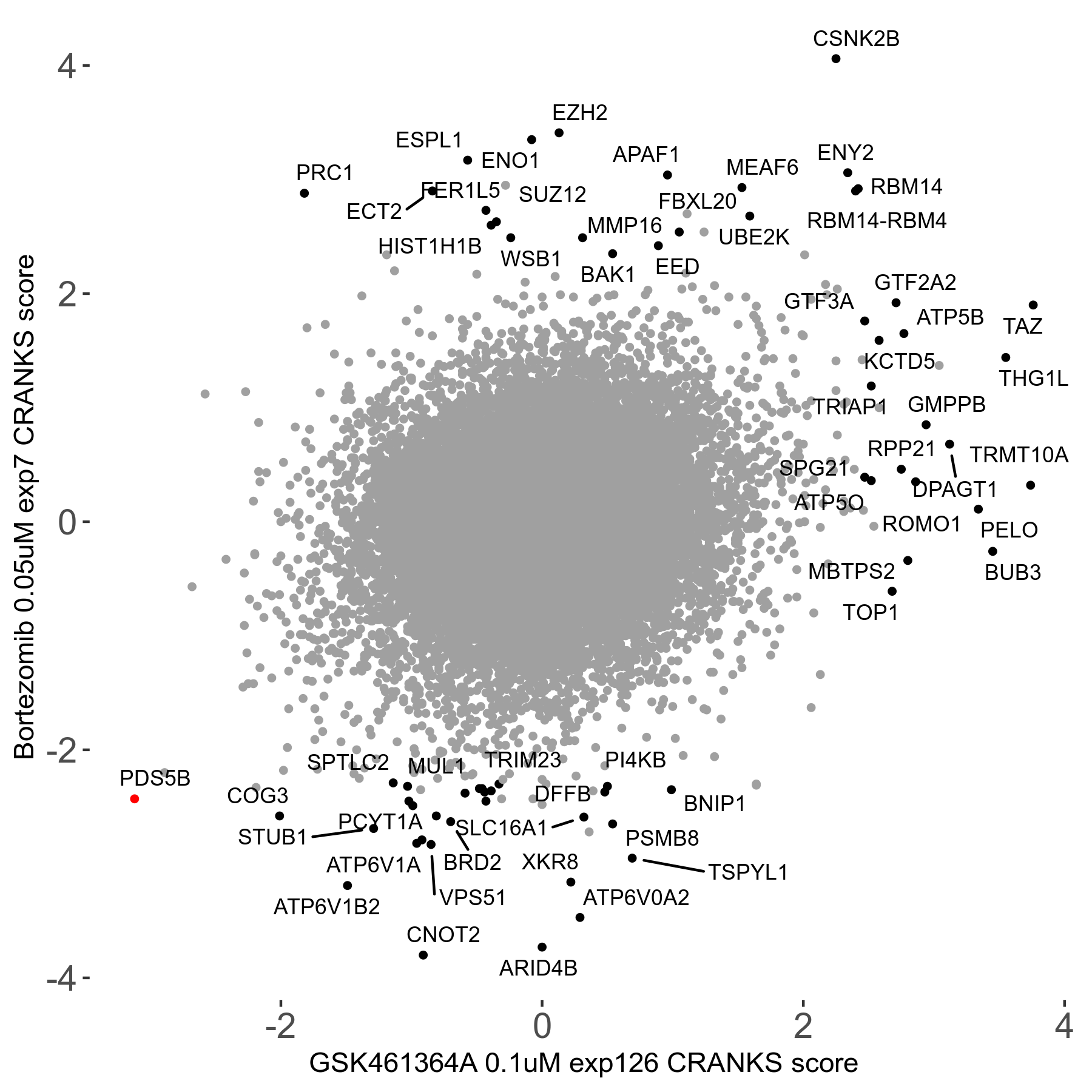
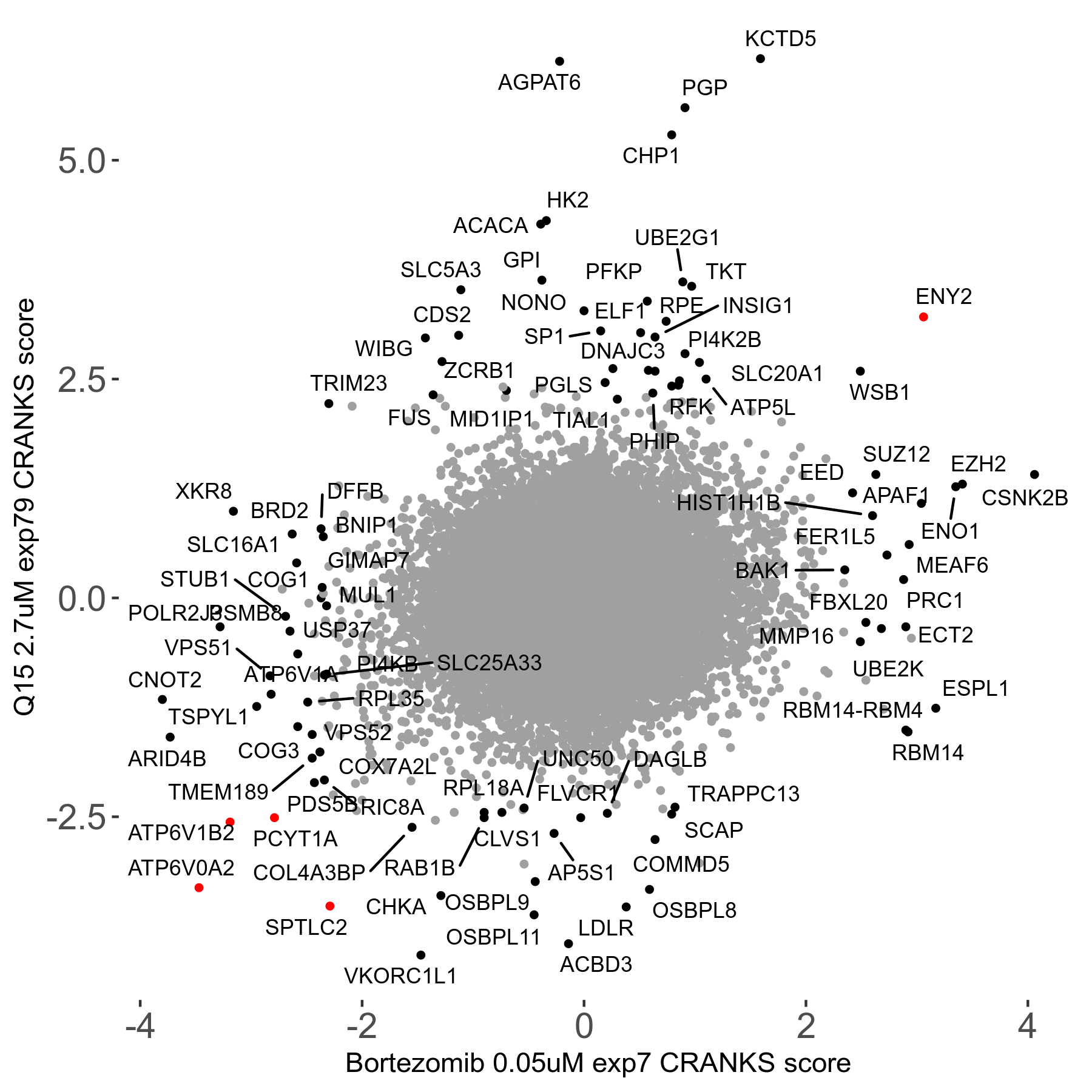
Bortezomib 0.05μM R00 exp7
Mechanism of Action
Inhibits 26S proteasome, binds reversibly to chymotrypsin-like catalytic sites
- Class / Subclass 1: Proteostasis / Proteasome Inhibitor
Technical Notes
Protein References
- PubChem Name: Bortezomib
- Synonyms: PS-341; Brotezamide; DPBA; LDP 341; MG 341; Radiciol; NSC 681239
- CAS #: 179324-69-7
- PubChem CID: 387447
- IUPAC: [(1R)-3-methyl-1-[[(2S)-3-phenyl-2-(pyrazine-2-carbonylamino)propanoyl]amino]butyl]boronic acid
- INCHI Name: InChI=1S/C19H25BN4O4/c1-13(2)10-17(20(27)28)24-18(25)15(11-14-6-4-3-5-7-14)23-19(26)16-12-21-8-9-22-16/h3-9,12-13,15,17,27-28H,10-11H2,1-2H3,(H,23,26)(H,24,25)/t15-,17-/m0/s1
- INCHI Key: GXJABQQUPOEUTA-RDJZCZTQSA-N
- Molecular Weight: 384.2
- Canonical SMILES: B(C(CC(C)C)NC(=O)C(CC1=CC=CC=C1)NC(=O)C2=NC=CN=C2)(O)O
- Isomeric SMILES: B([C@H](CC(C)C)NC(=O)[C@H](CC1=CC=CC=C1)NC(=O)C2=NC=CN=C2)(O)O
- Molecular Formula: C19H25BN4O4
Protein Supplier
- Supplier Name: LC Laboratories
- Catalog #: B-1408
- Lot #: BBZ-113
Protein Characterization
- HRMS (ESI-TOF) m/z: [M+H-H2O]+ Calcd for C19H24BN4O3 367.1936; found 367.1966
Dose Response Curve
- Platform ID: Bortezomib
- Min: -9.7617; Max: 99.4238

| IC | Concentration (µM) |
|---|---|
| IC10 | N/A |
| IC20 | 0.0070 |
| IC30 | 0.0072 |
| IC40 | 0.0073 |
| IC50 | 0.0074 |
| IC60 | N/A |
| IC70 | N/A |
| IC80 | N/A |
| IC90 | N/A |
Screen Summary
- Round: 00
- Dose: 50nM
- Days of incubation: 8
- Doublings: 0.3
- Numbers of reads: 12274814
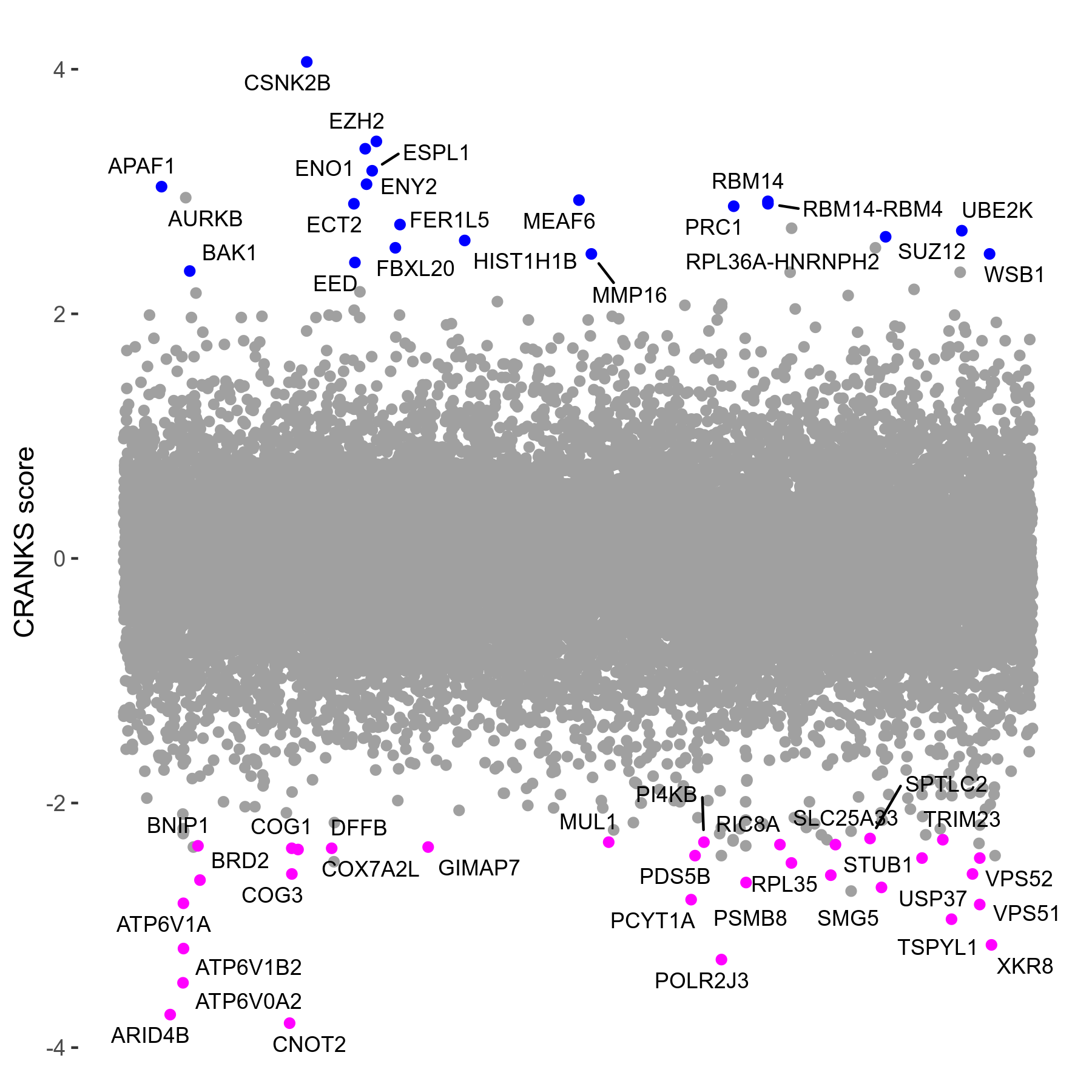
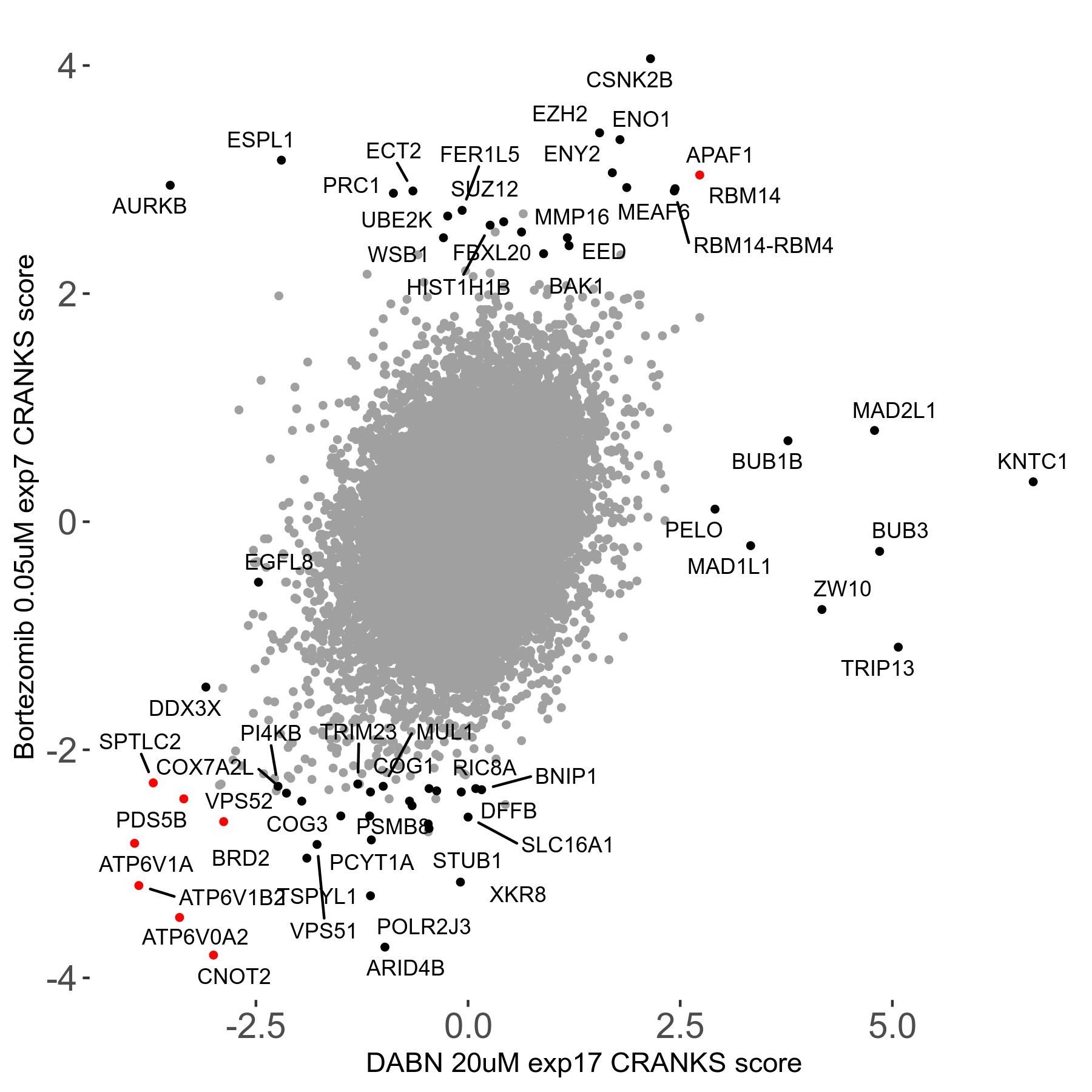
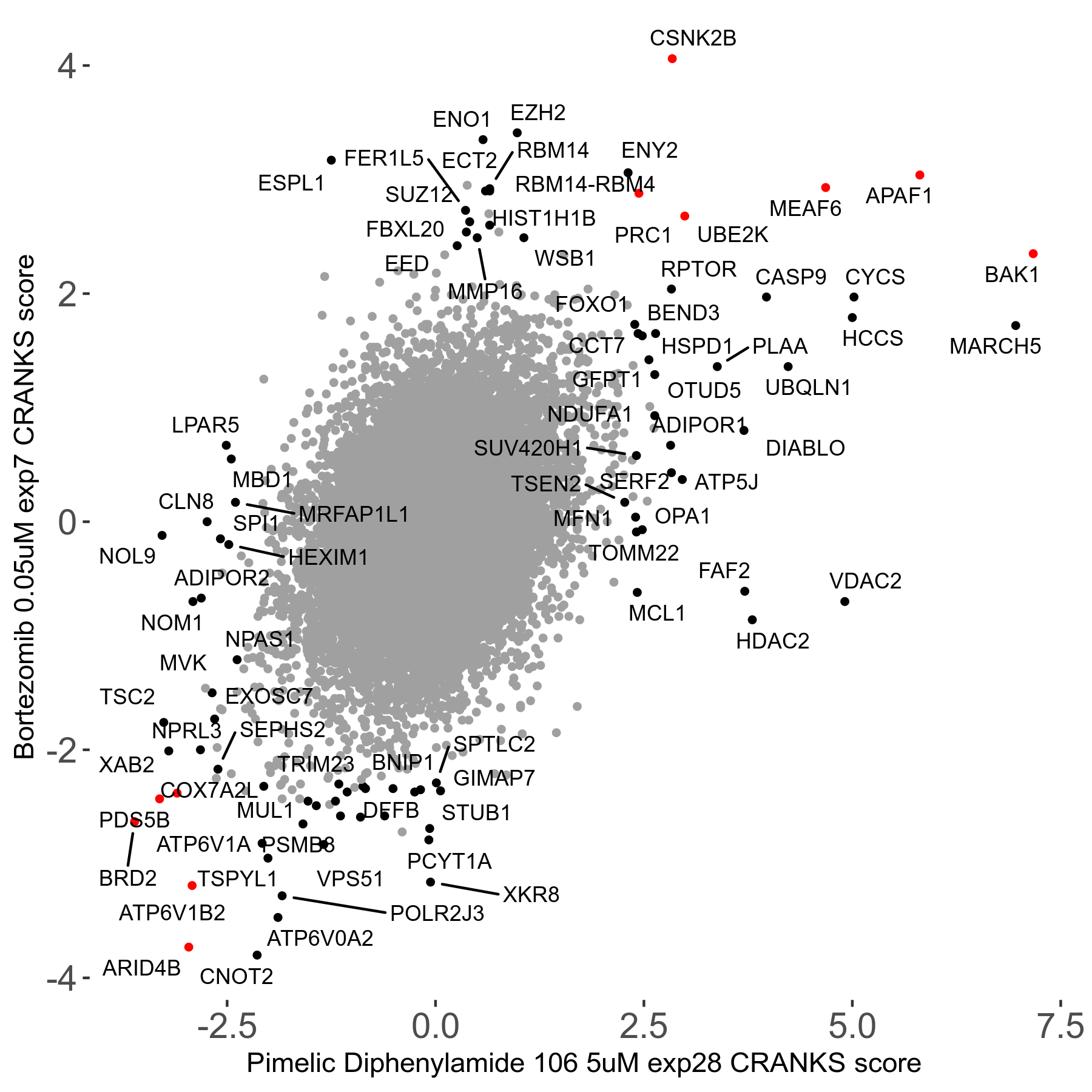
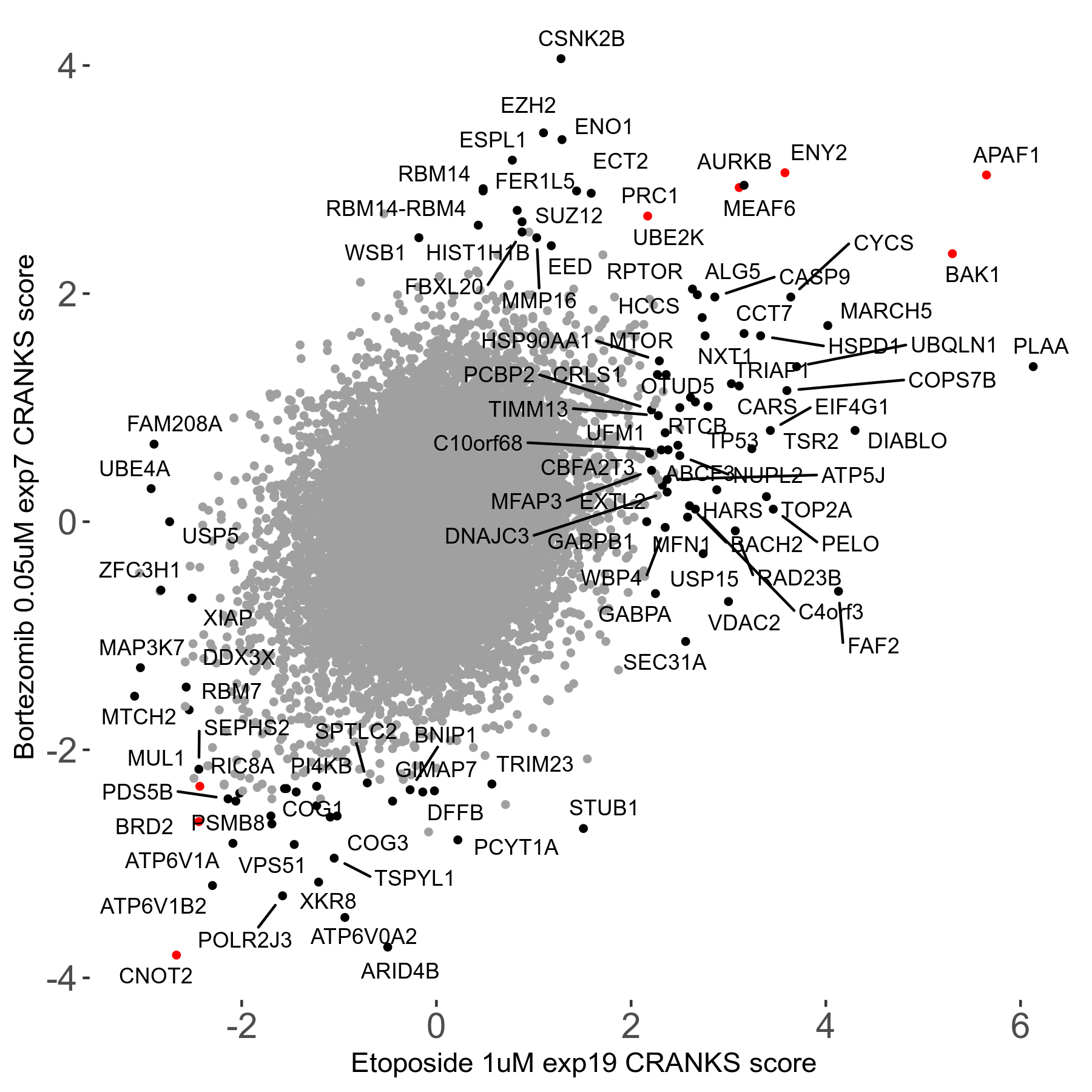
Screen Results
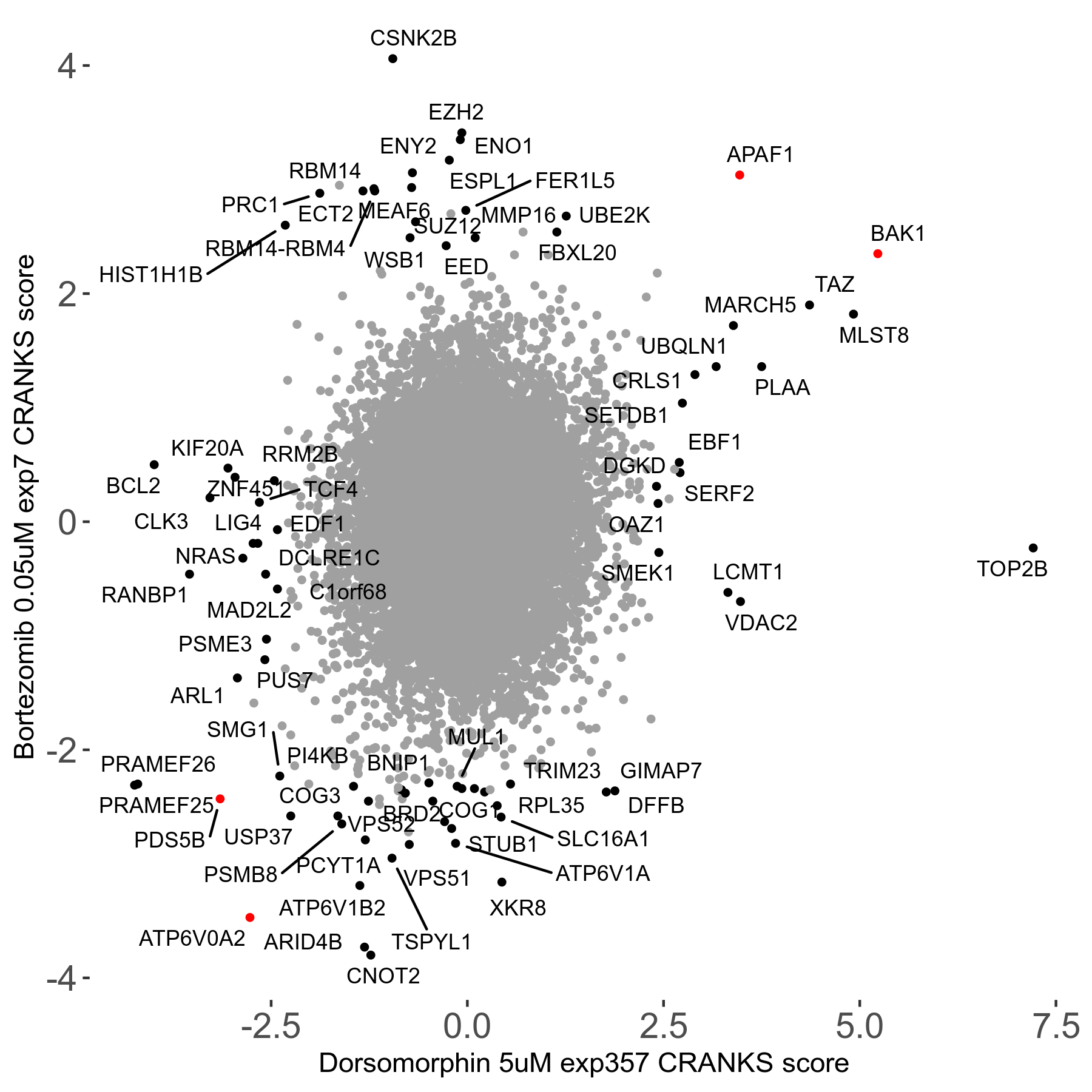
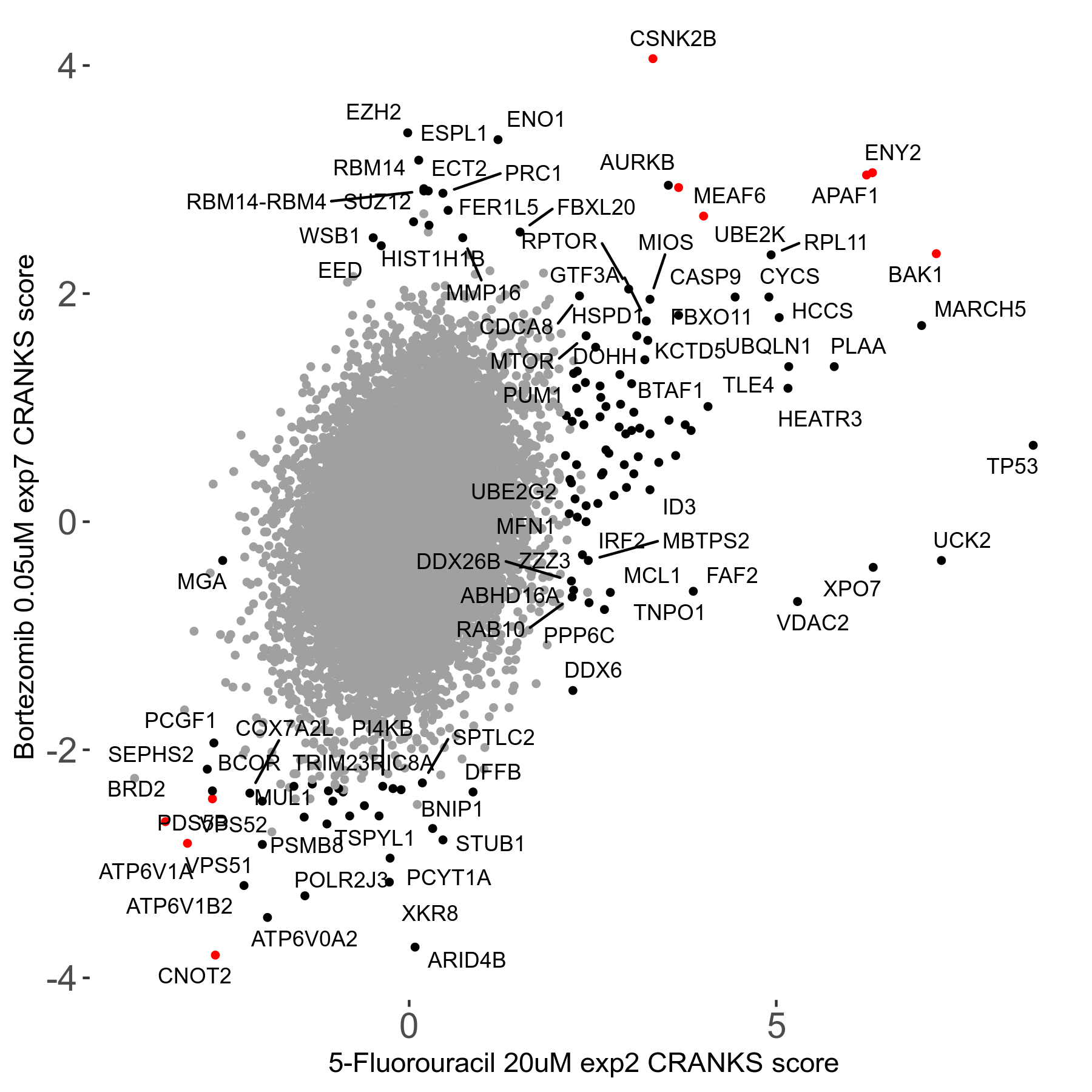
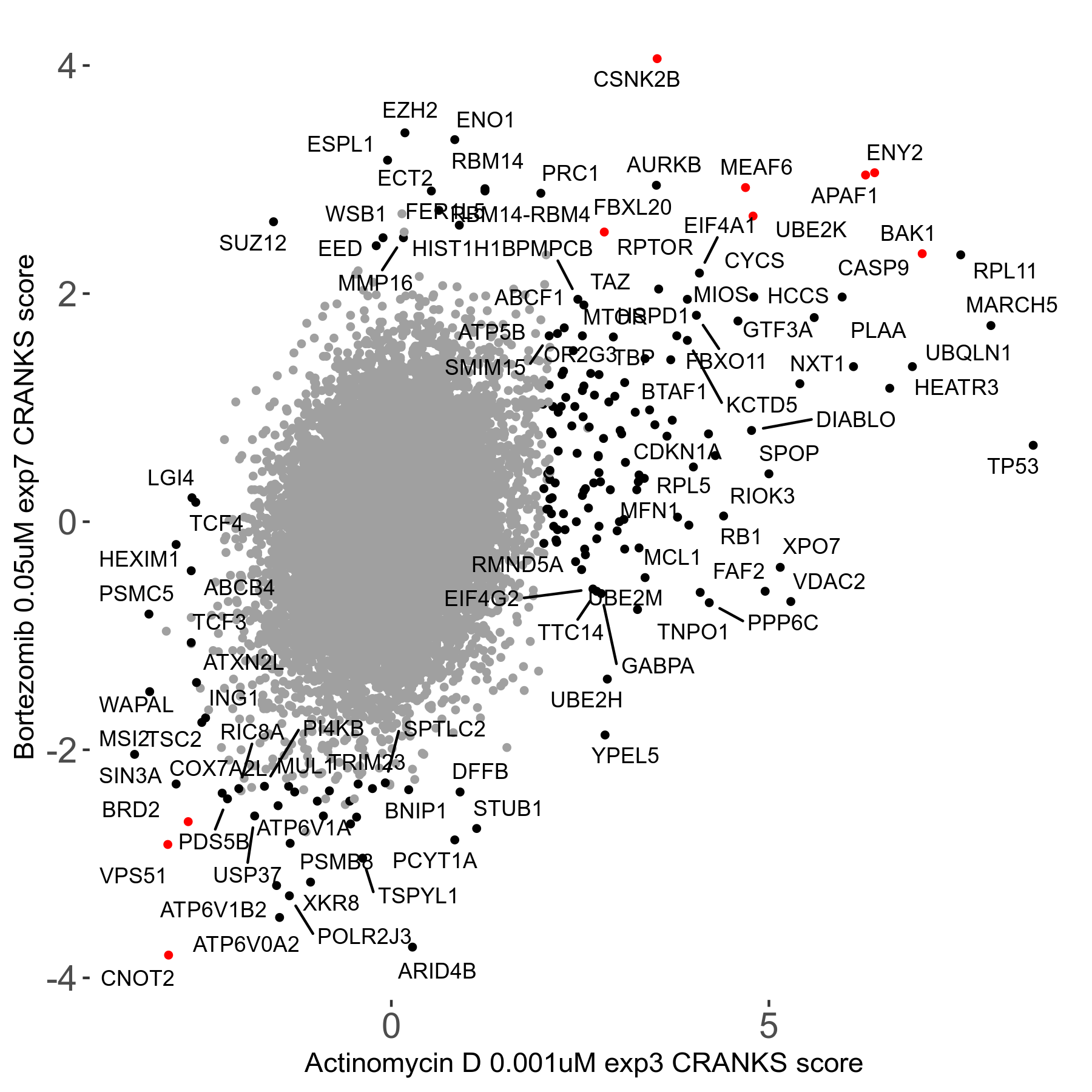
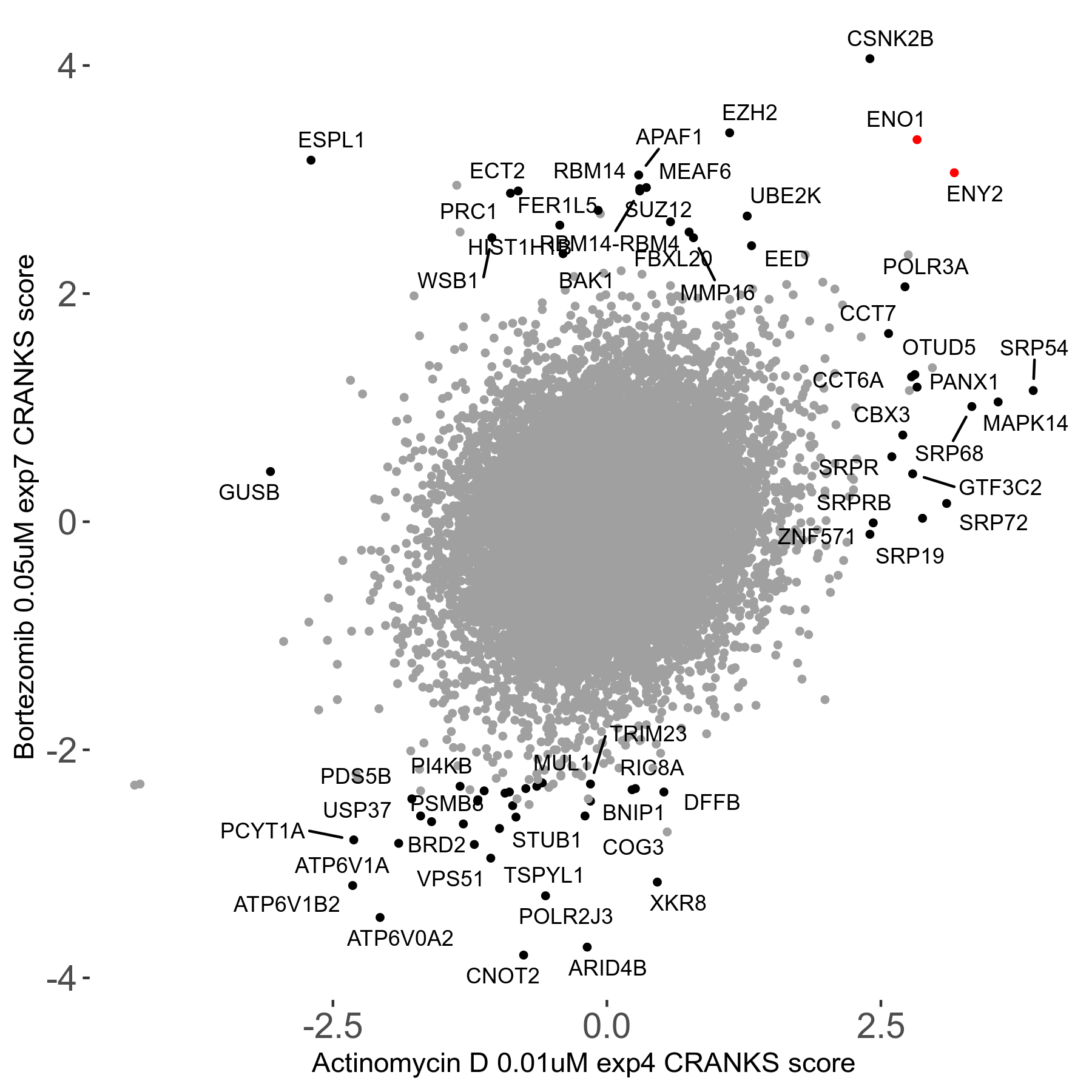
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 31/20 | Scores |