Table of Contents
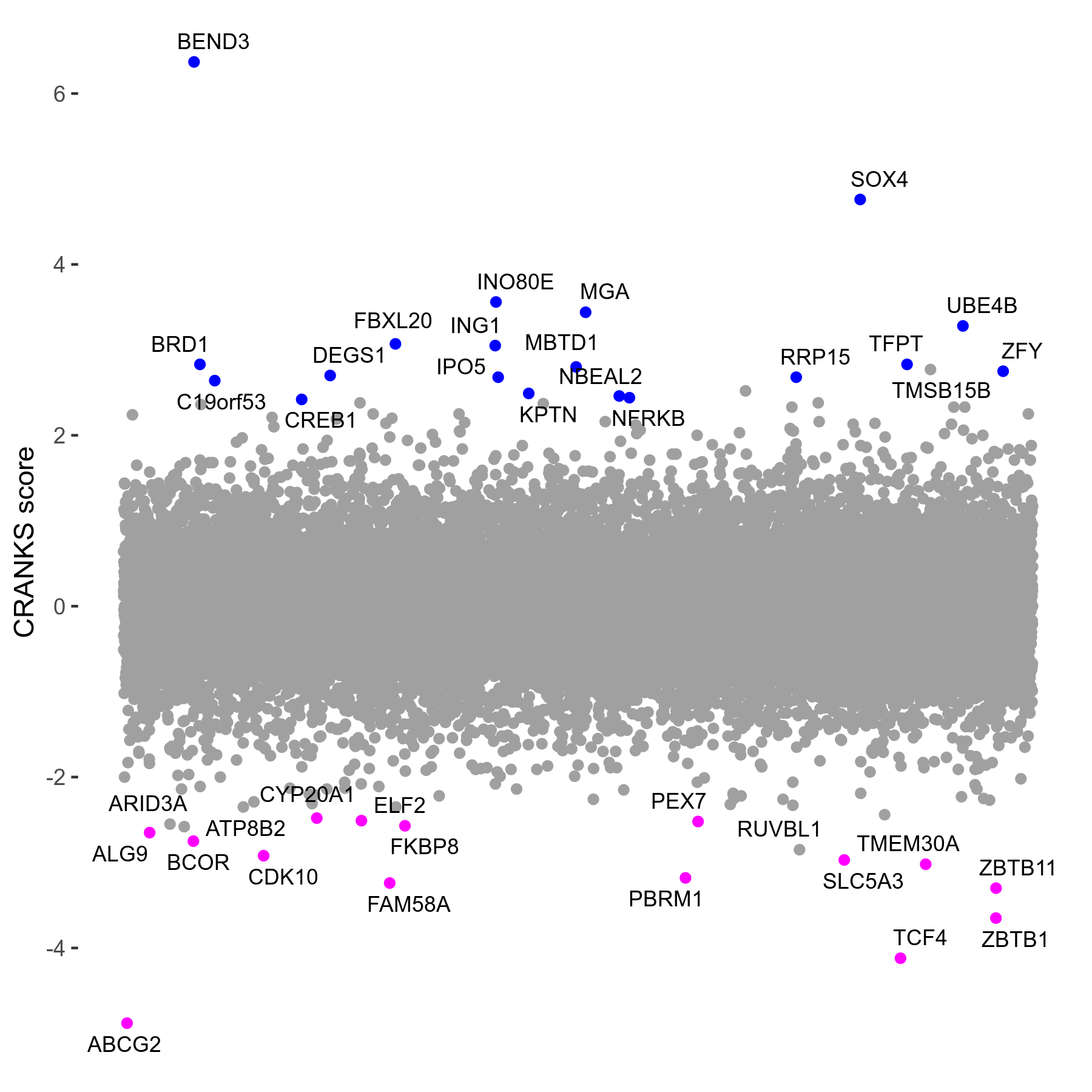
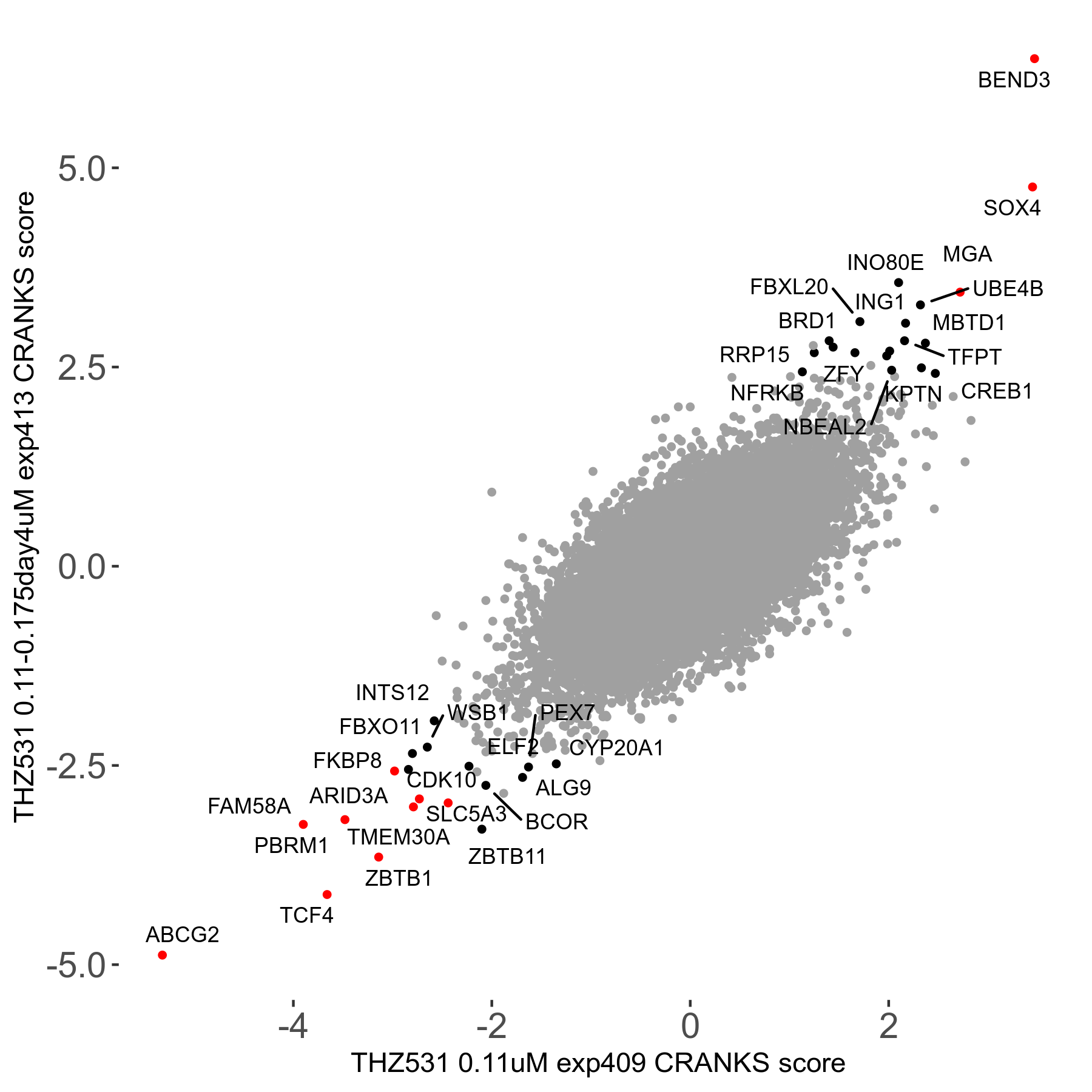
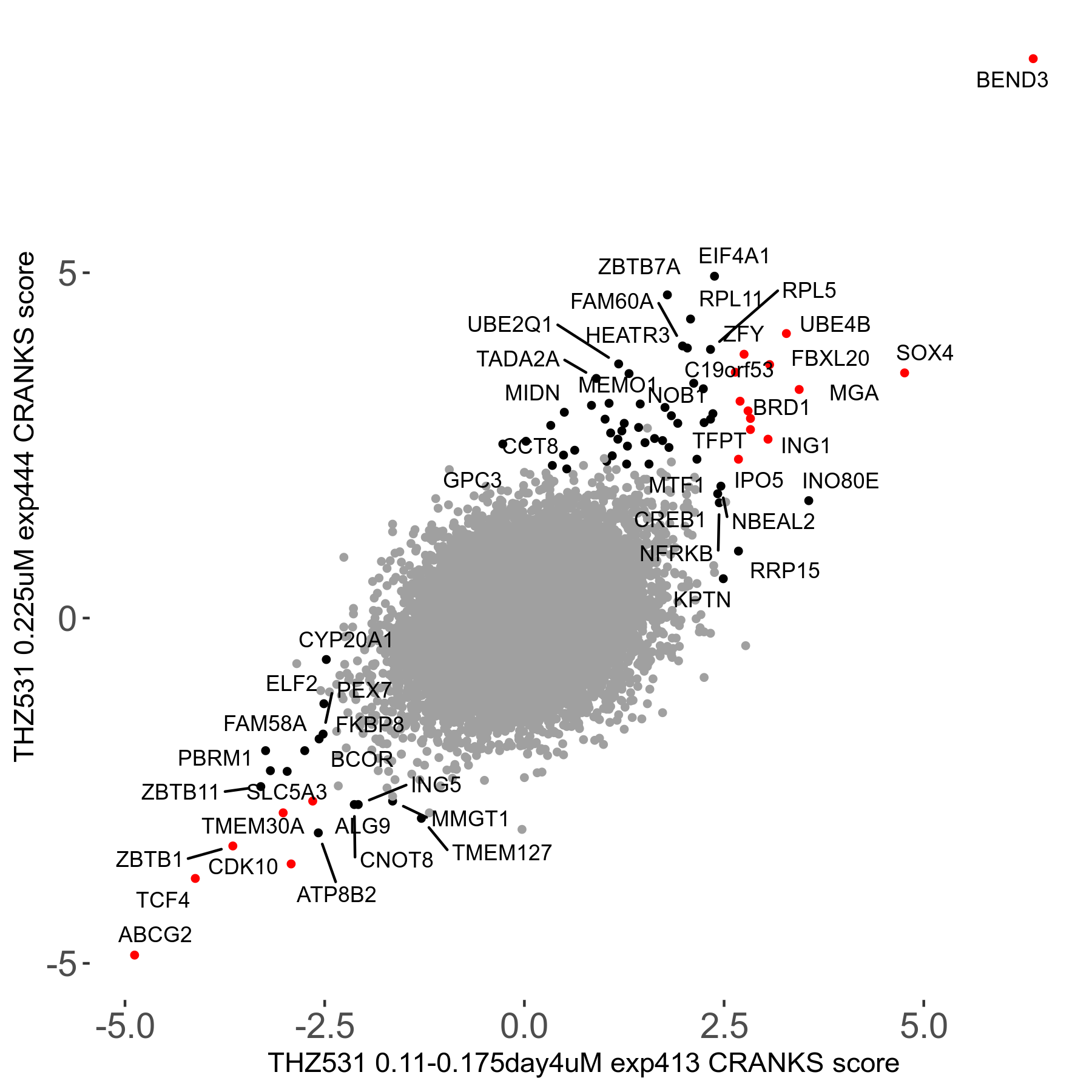
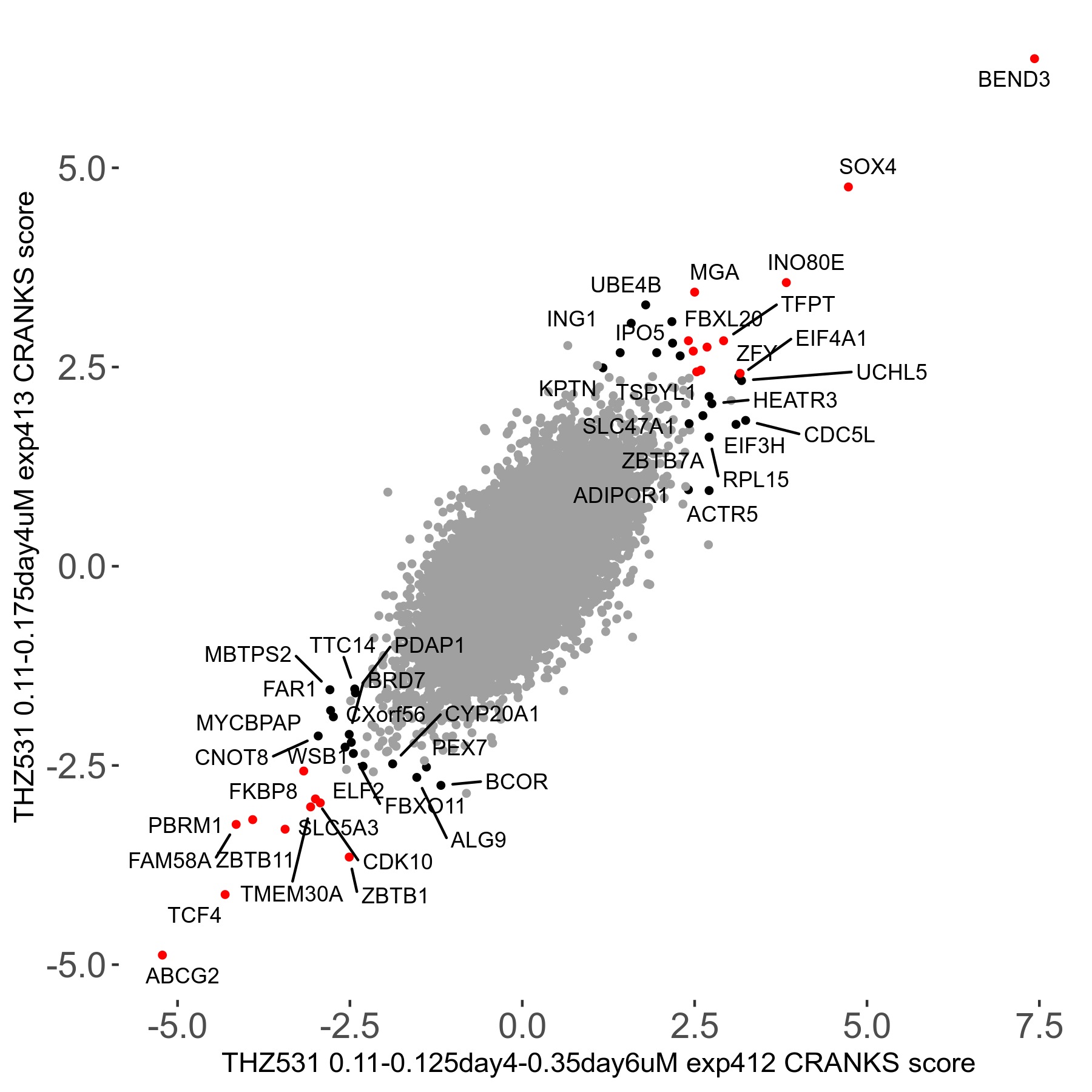
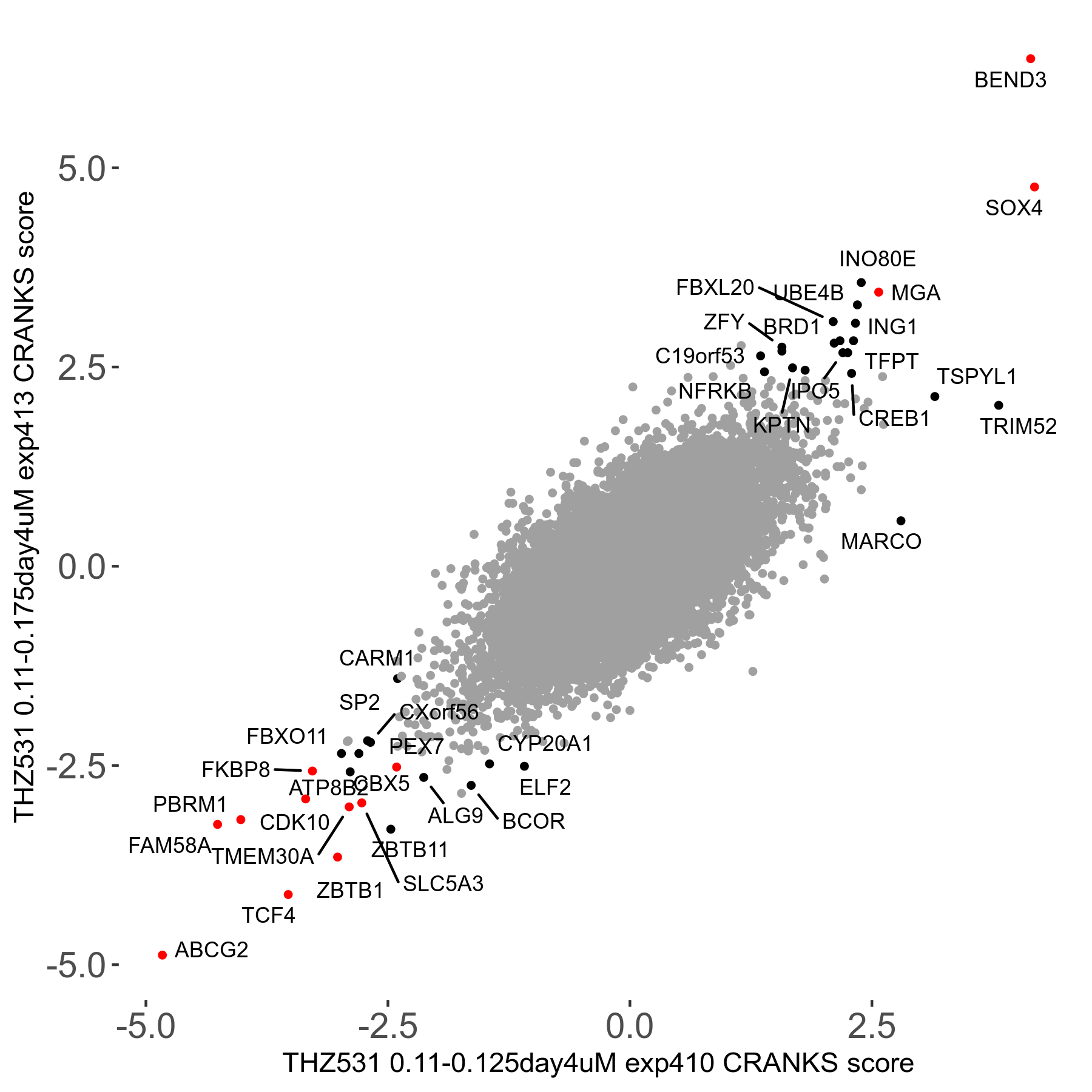
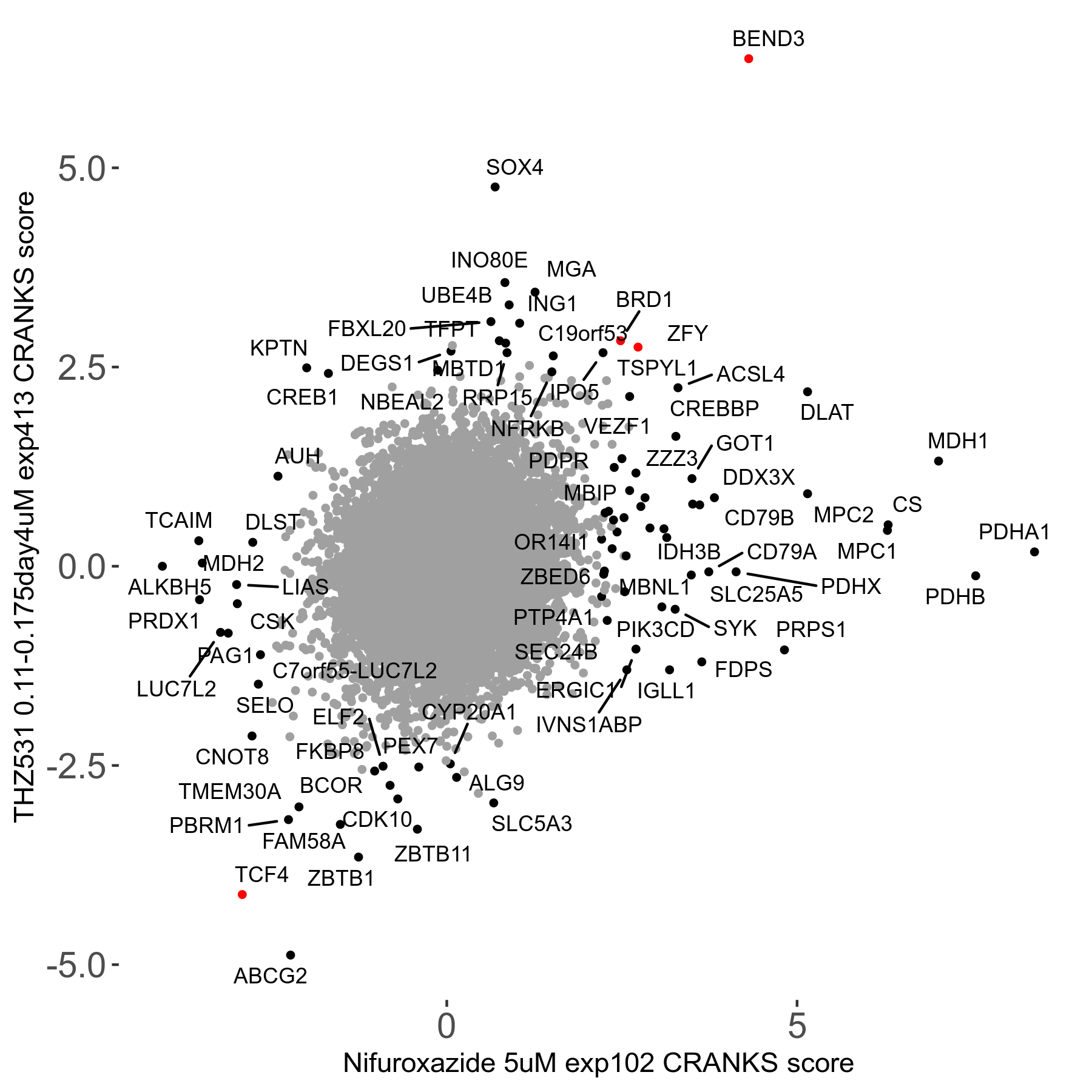
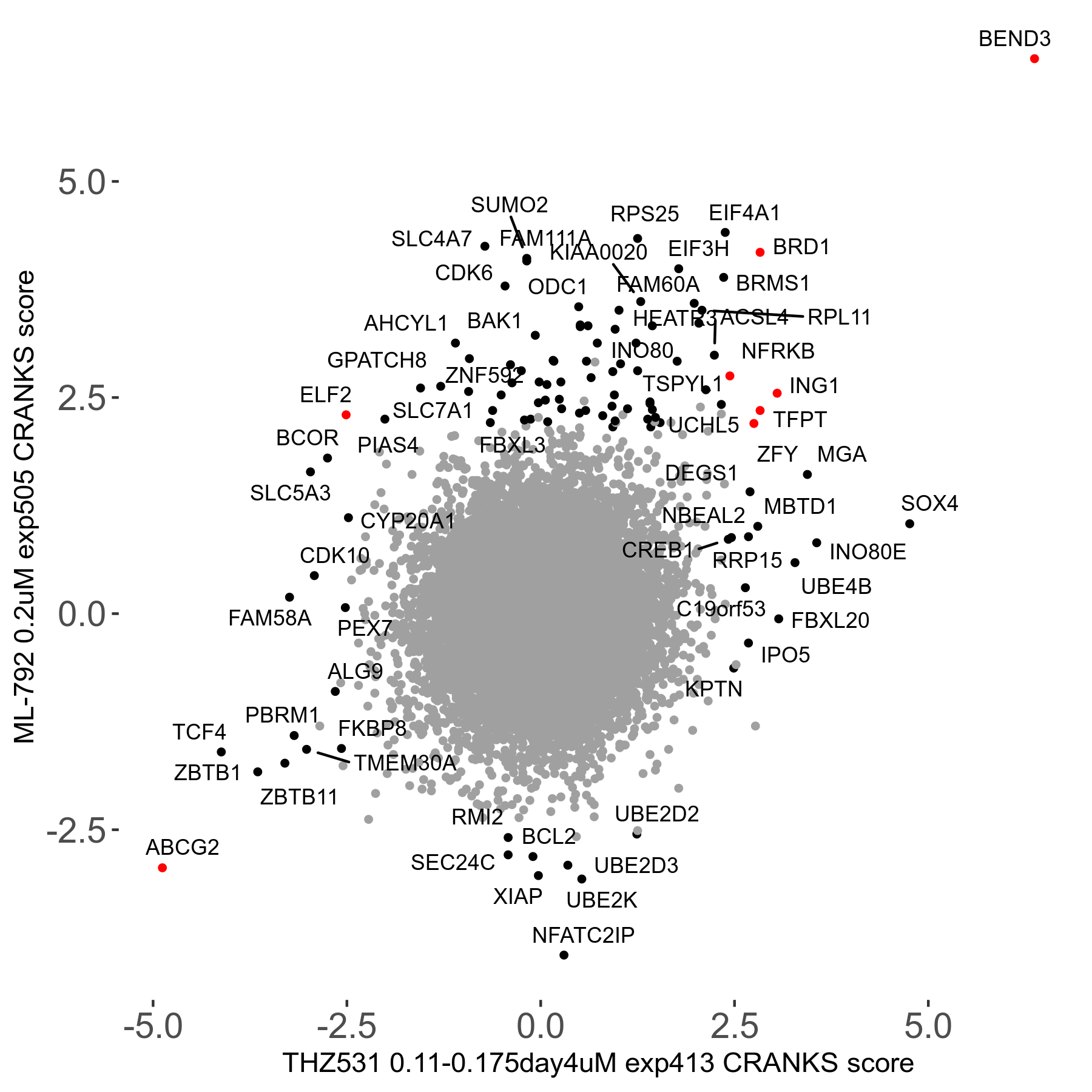
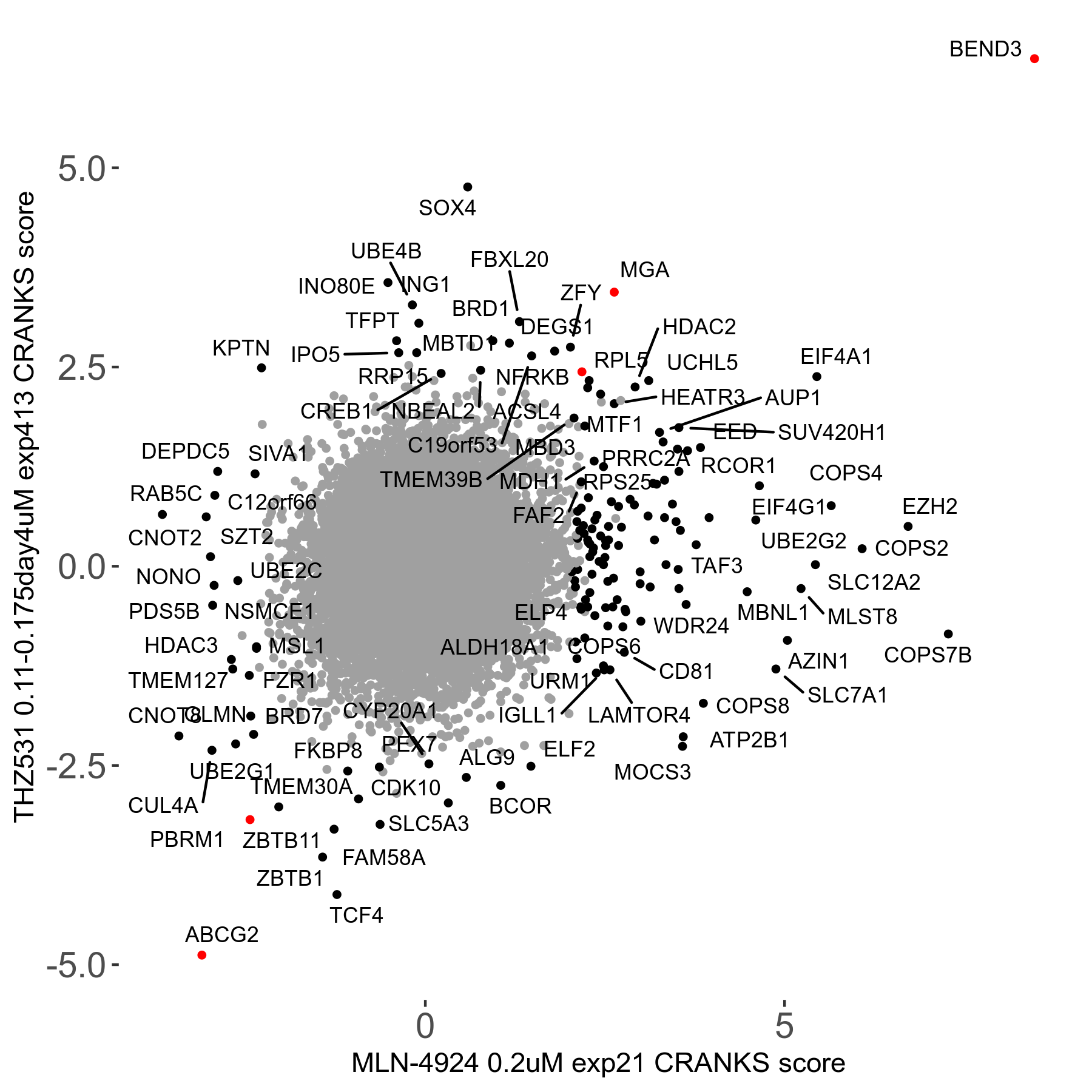
THZ531 0.11 to 0.175μM on day4 R07 exp413
Mechanism of Action
Inhibits CDK12 and CDK13, forms covalent adduct, more selective analog of THZ1, blocks phosphorylation of the C-terminal domain of RNA polymerase II
- Class / Subclass 1: Gene Regulation / Transcription Inhibitor
- Class / Subclass 2: Signal Transduction / Kinase Inhibitor
Technical Notes
Protein References
- PubChem Name: (E)-N-[4-[(3R)-3-[[5-Chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidine-1-carbonyl]phenyl]-4-(dimethylamino)but-2-enamide
- Synonyms: N/A
- CAS #: 1702809-17-3
- PubChem CID: 118025540
- IUPAC: (E)-N-[4-[(3R)-3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidine-1-carbonyl]phenyl]-4-(dimethylamino)but-2-enamide
- INCHI Name: InChI=1S/C30H32ClN7O2/c1-37(2)15-6-10-27(39)34-21-13-11-20(12-14-21)29(40)38-16-5-7-22(19-38)35-30-33-18-25(31)28(36-30)24-17-32-26-9-4-3-8-23(24)26/h3-4,6,8-14,17-18,22,32H,5,7,15-16,19H2,1-2H3,(H,34,39)(H,33,35,36)/b10-6+/t22-/m1/s1
- INCHI Key: RUBYHLPRZRMTJO-MOVYNIQHSA-N
- Molecular Weight: 558.1
- Canonical SMILES: CN(C)CC=CC(=O)NC1=CC=C(C=C1)C(=O)N2CCCC(C2)NC3=NC=C(C(=N3)C4=CNC5=CC=CC=C54)Cl
- Isomeric SMILES: CN(C)C/C=C/C(=O)NC1=CC=C(C=C1)C(=O)N2CCC[C@H](C2)NC3=NC=C(C(=N3)C4=CNC5=CC=CC=C54)Cl
- Molecular Formula: C30H32ClN7O2
Protein Supplier
- Supplier Name: Med Chem Express
- Catalog #: HY-103618
- Lot #: 28264
Protein Characterization
- HRMS (ESI-TOF) m/z: (M+H)+ Calcd for C30H32ClN7O2 558.23788; found 558.23943
Dose Response Curve
- Platform ID: THZ531
- Min: -7.6335; Max: 95.3893

| IC | Concentration (µM) |
|---|---|
| IC10 | N/A |
| IC20 | 0.0546 |
| IC30 | 0.0716 |
| IC40 | 0.0910 |
| IC50 | 0.1150 |
| IC60 | N/A |
| IC70 | N/A |
| IC80 | N/A |
| IC90 | N/A |
Screen Summary
- Round: 07
- Dose: 0.11-0.175µM
- Days of incubation: 8
- Doublings: 5.8
- Numbers of reads: 15770666
Screen Results
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 15/19 | Scores |