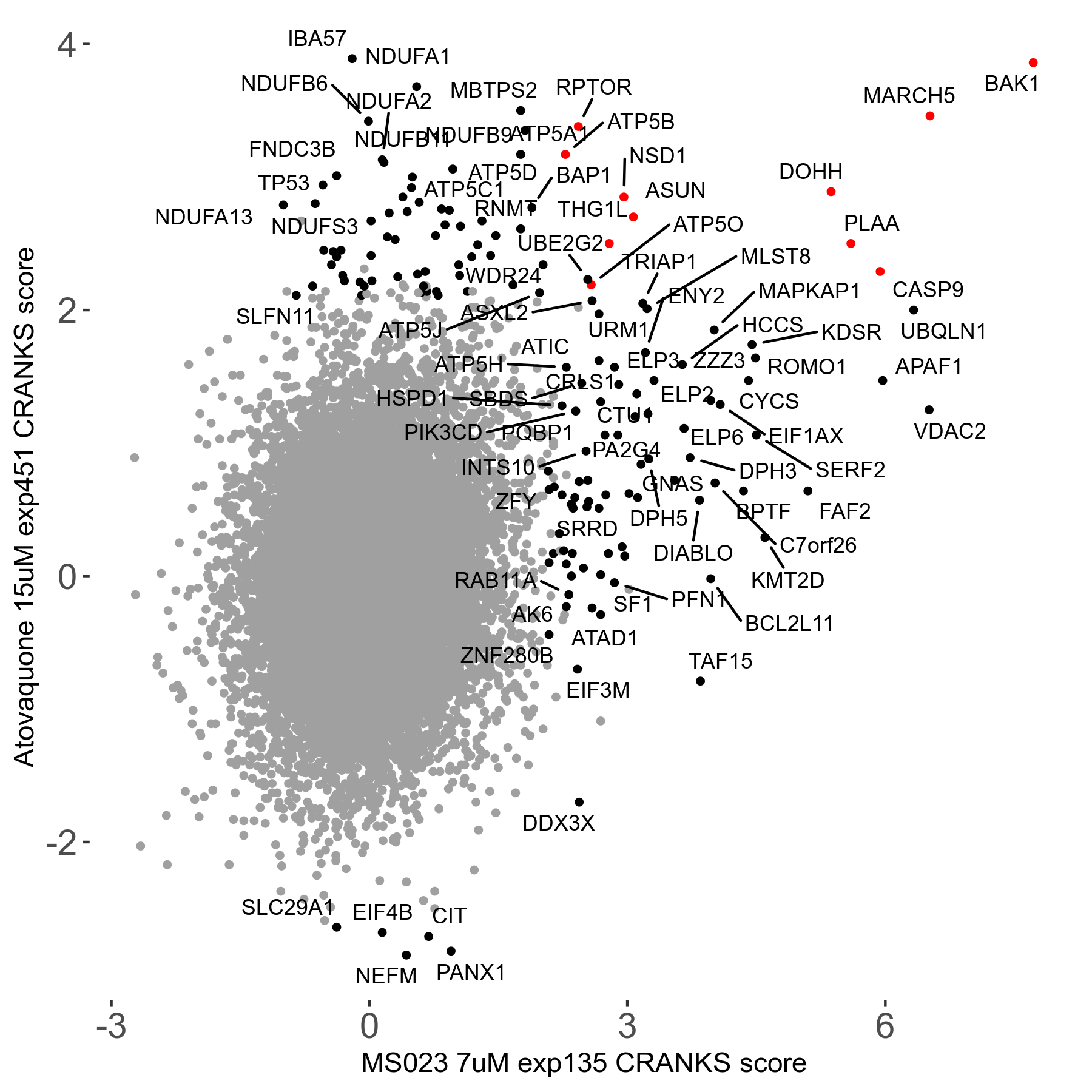
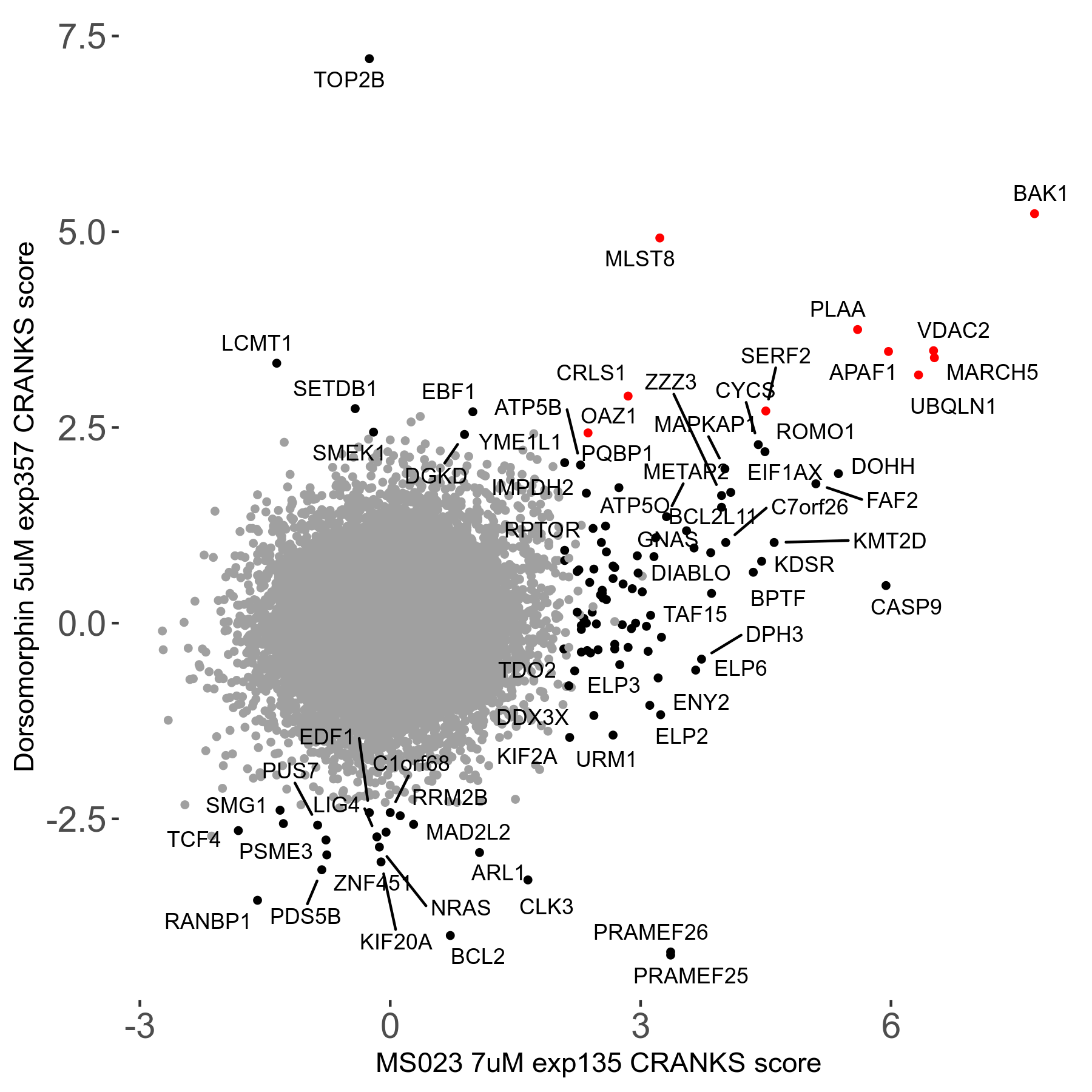
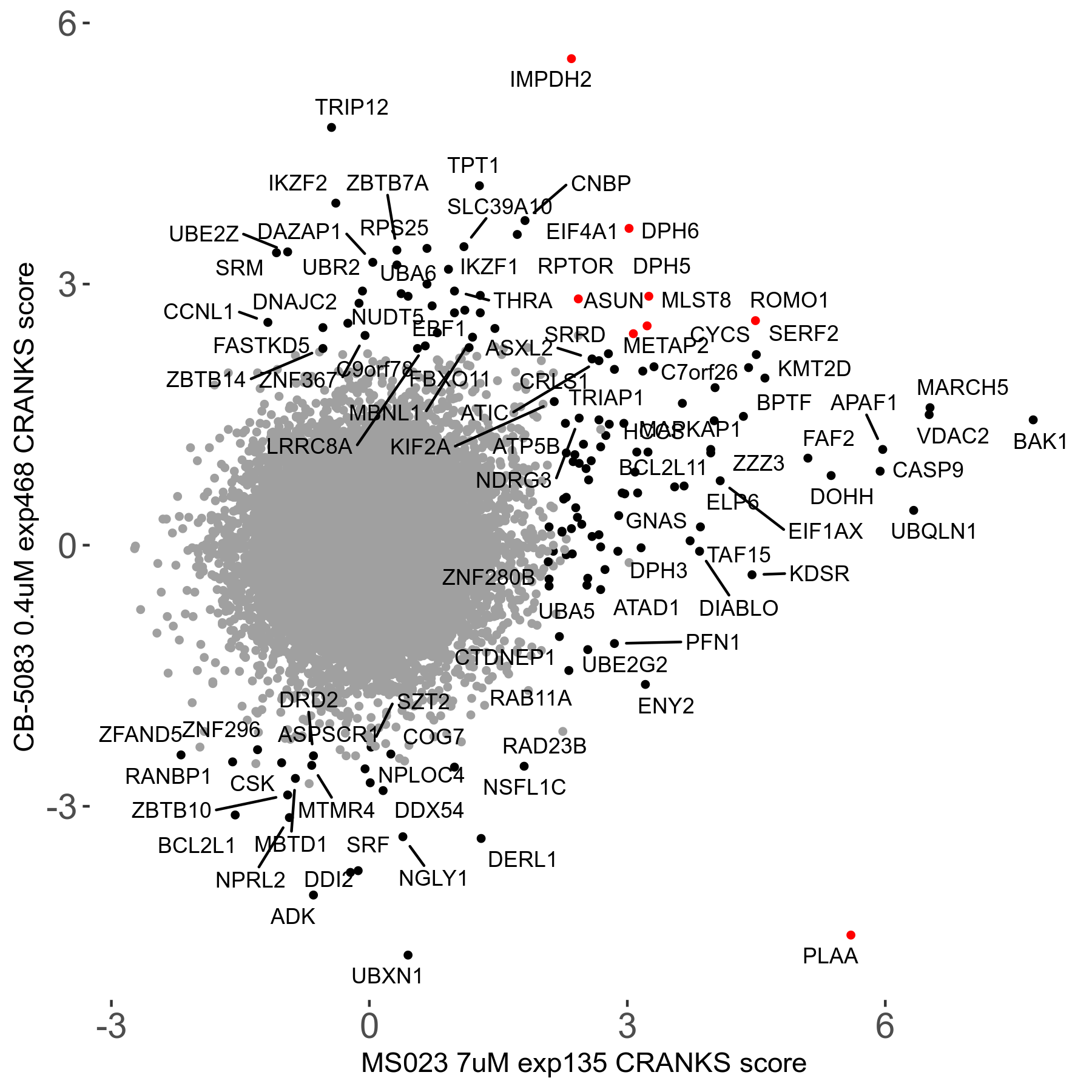
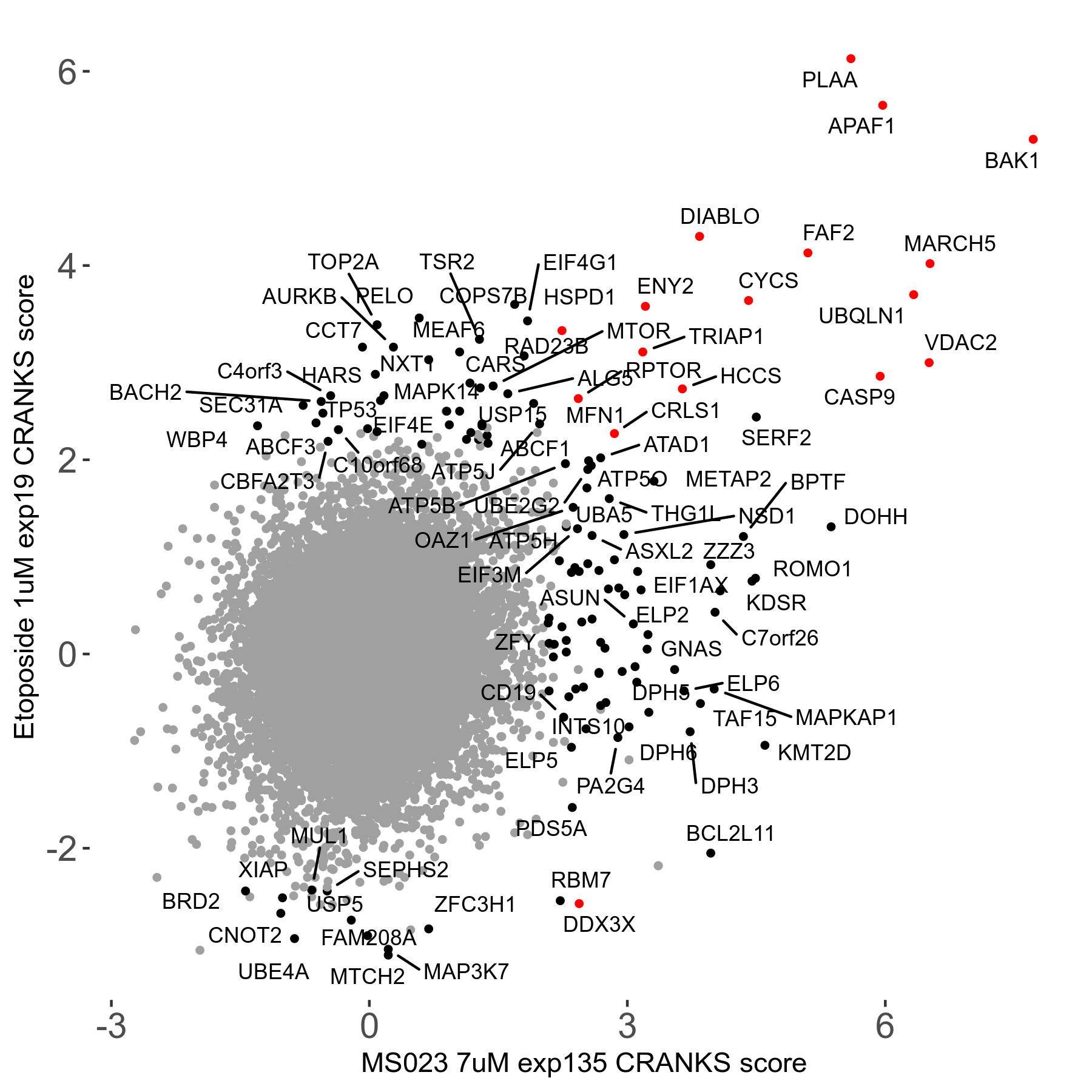
Table of Contents
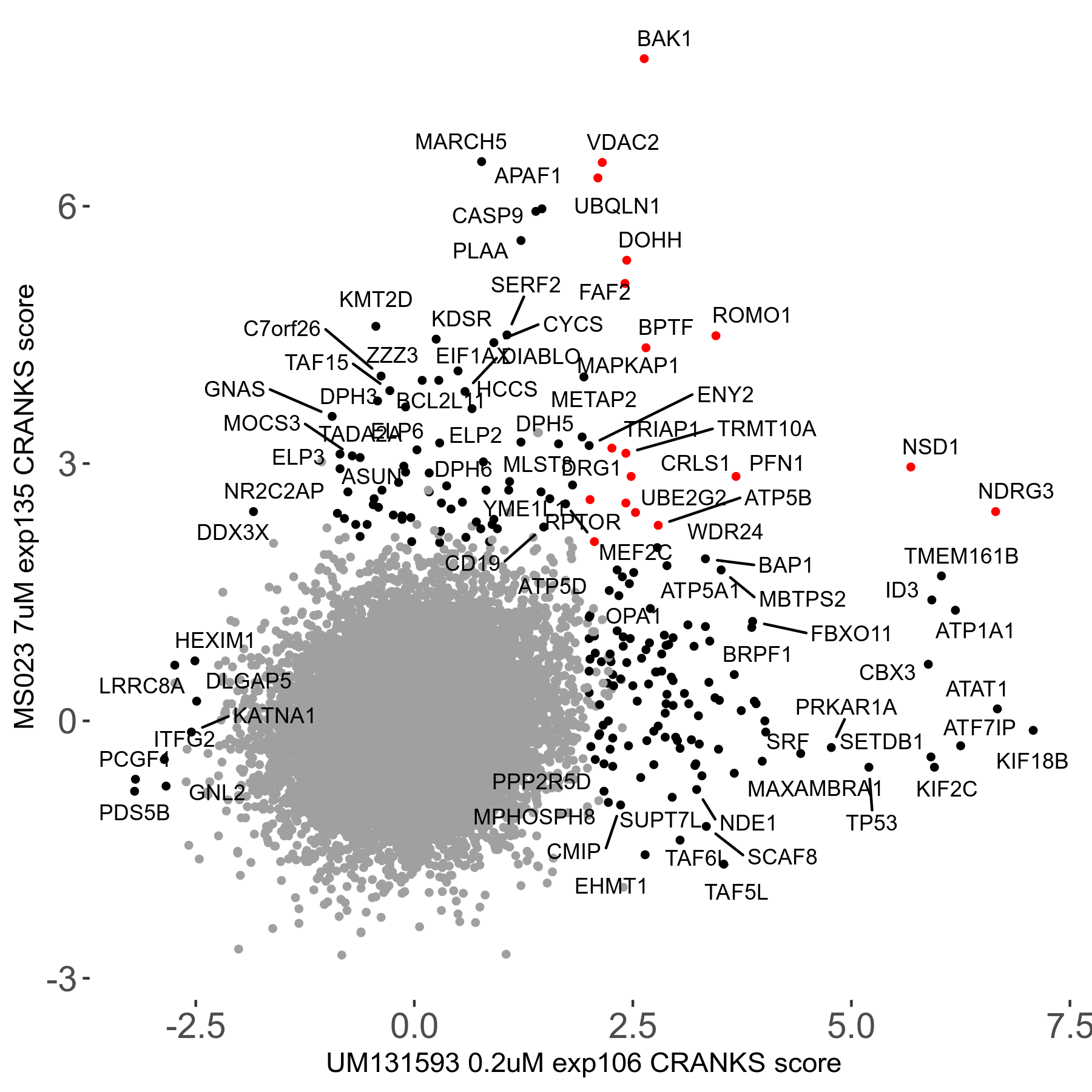
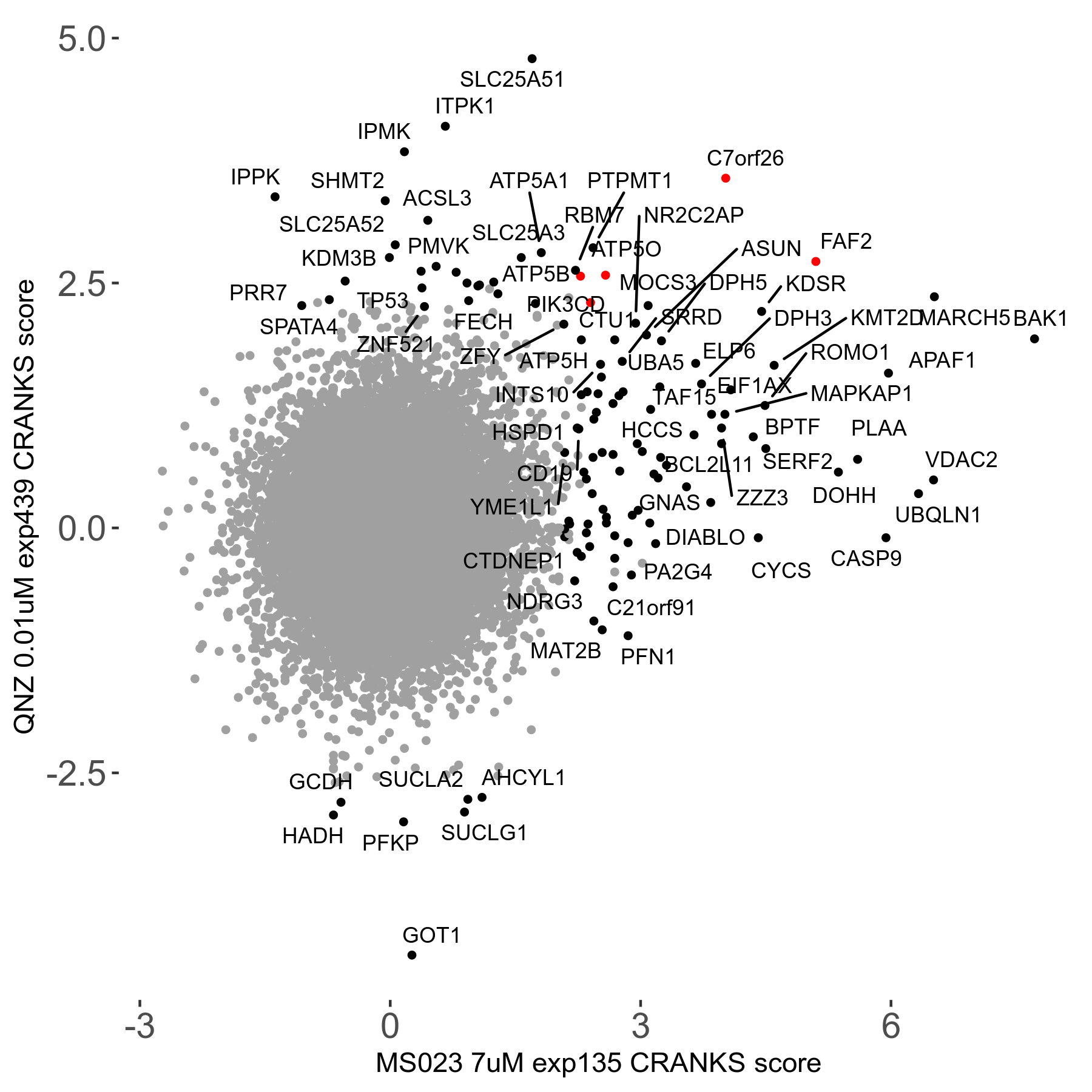
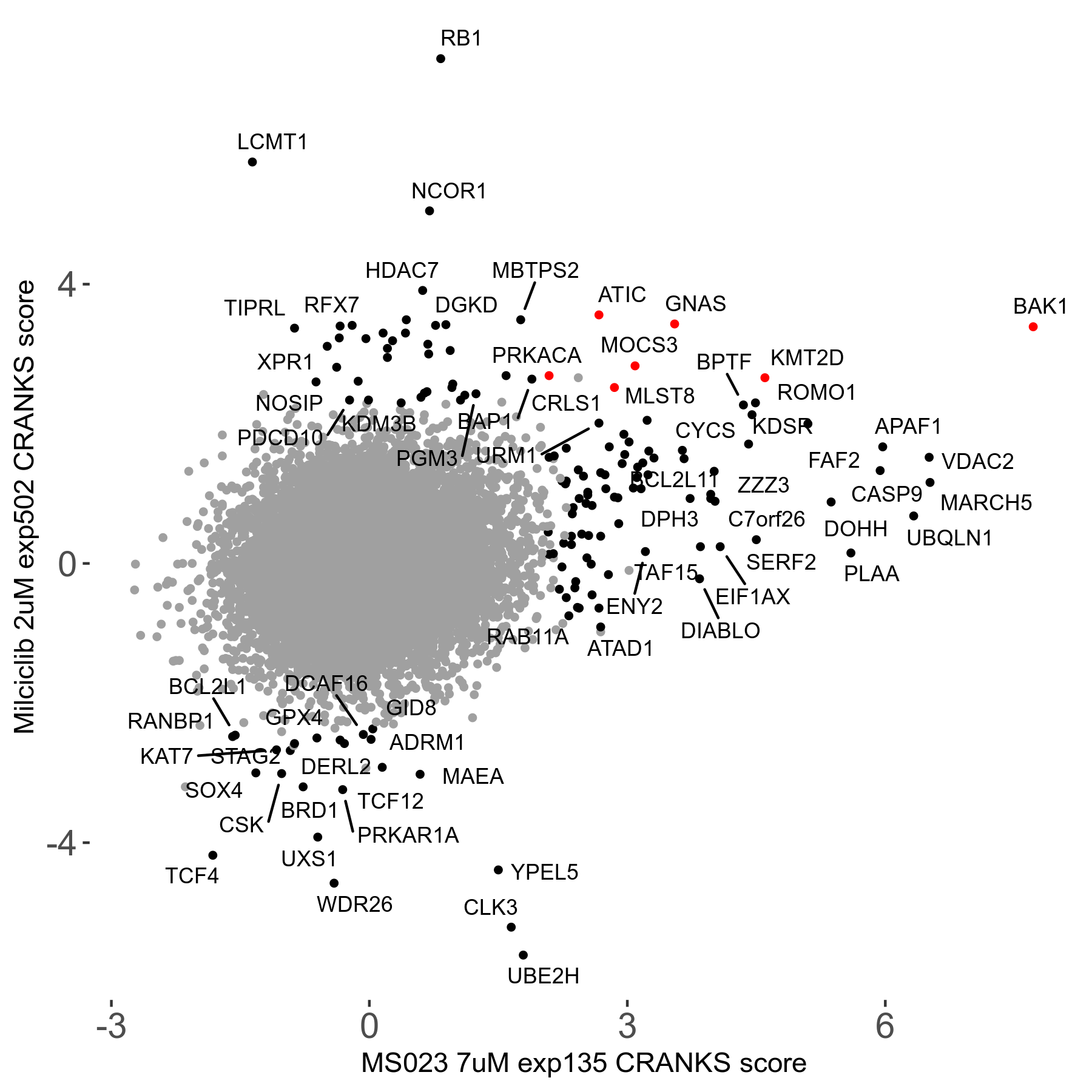
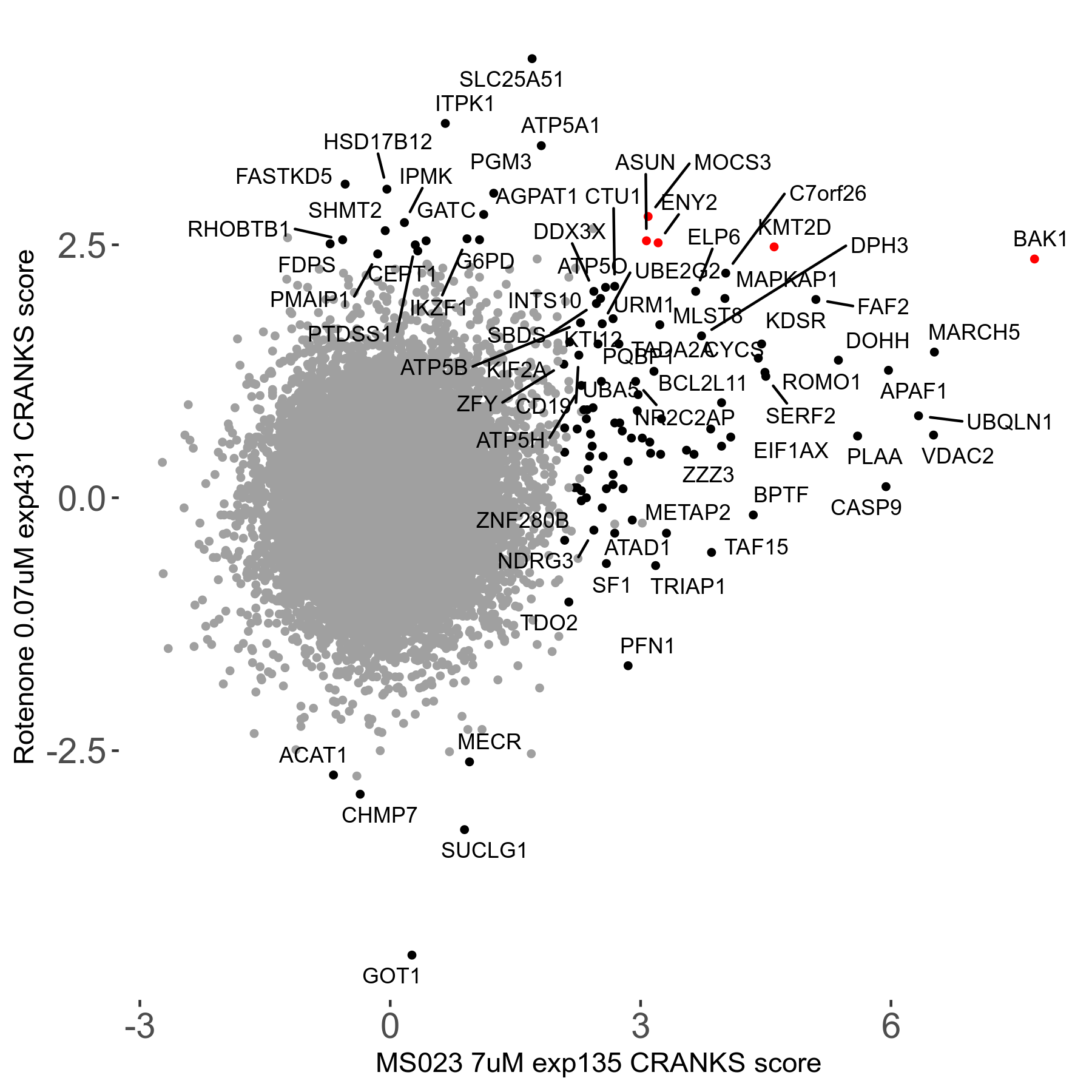
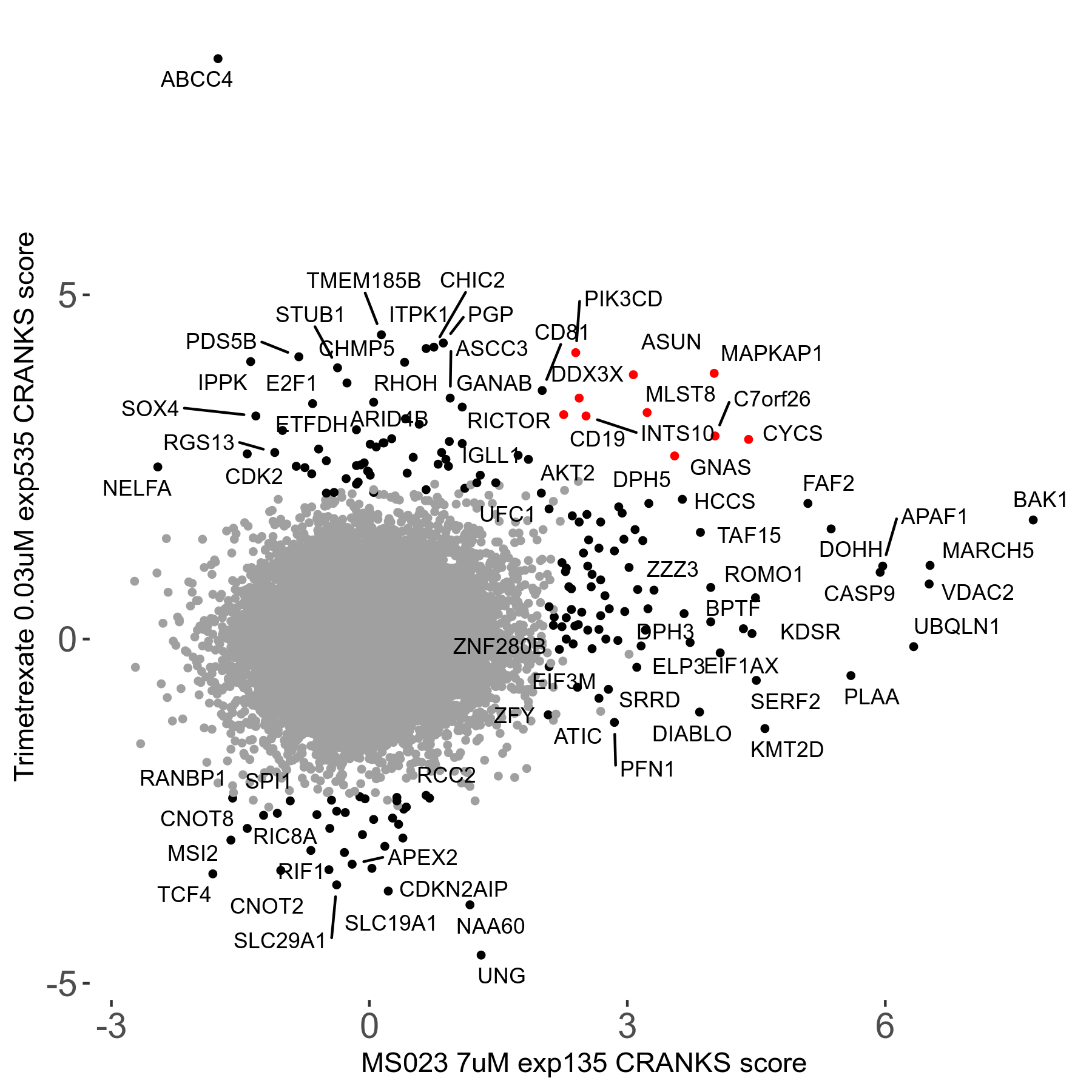
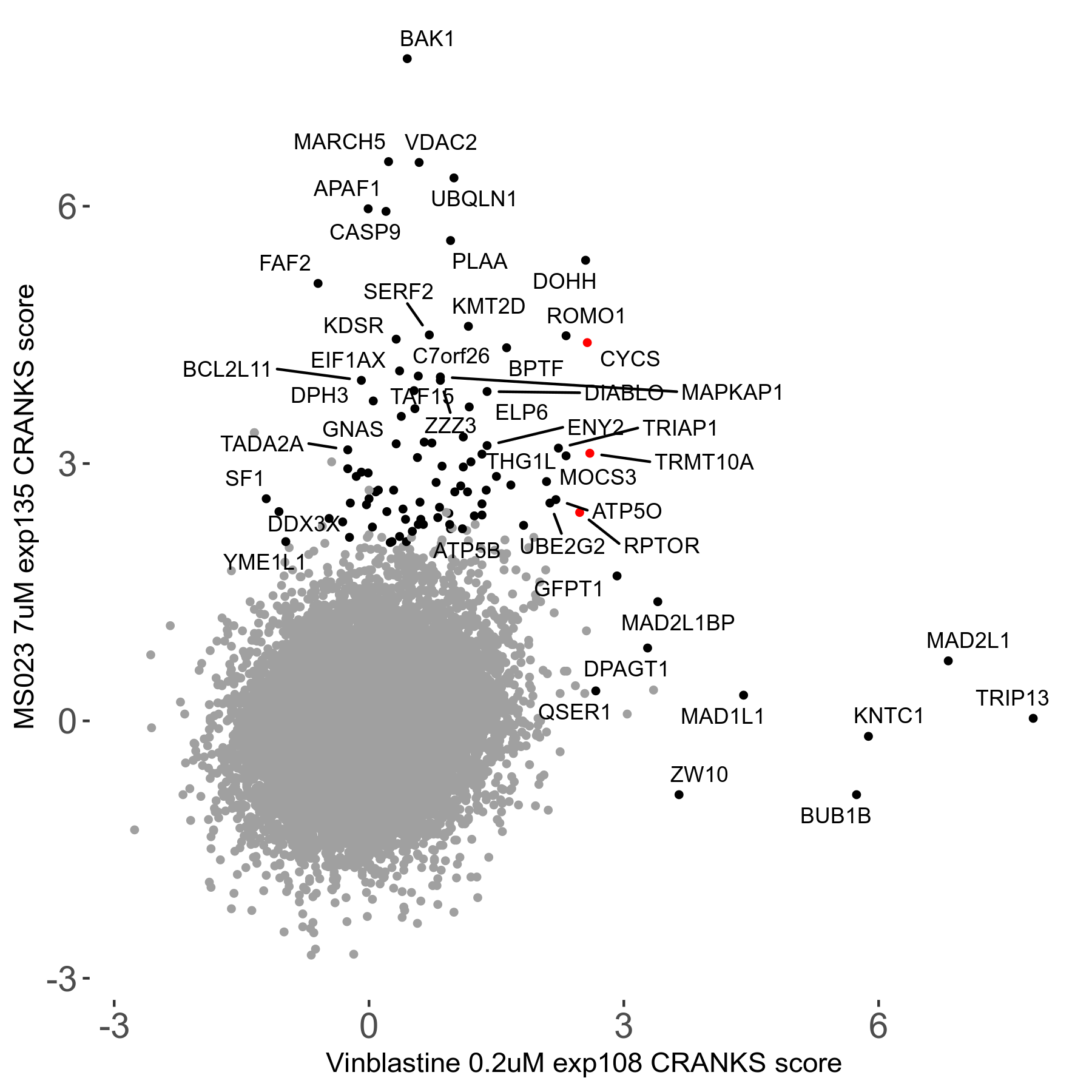
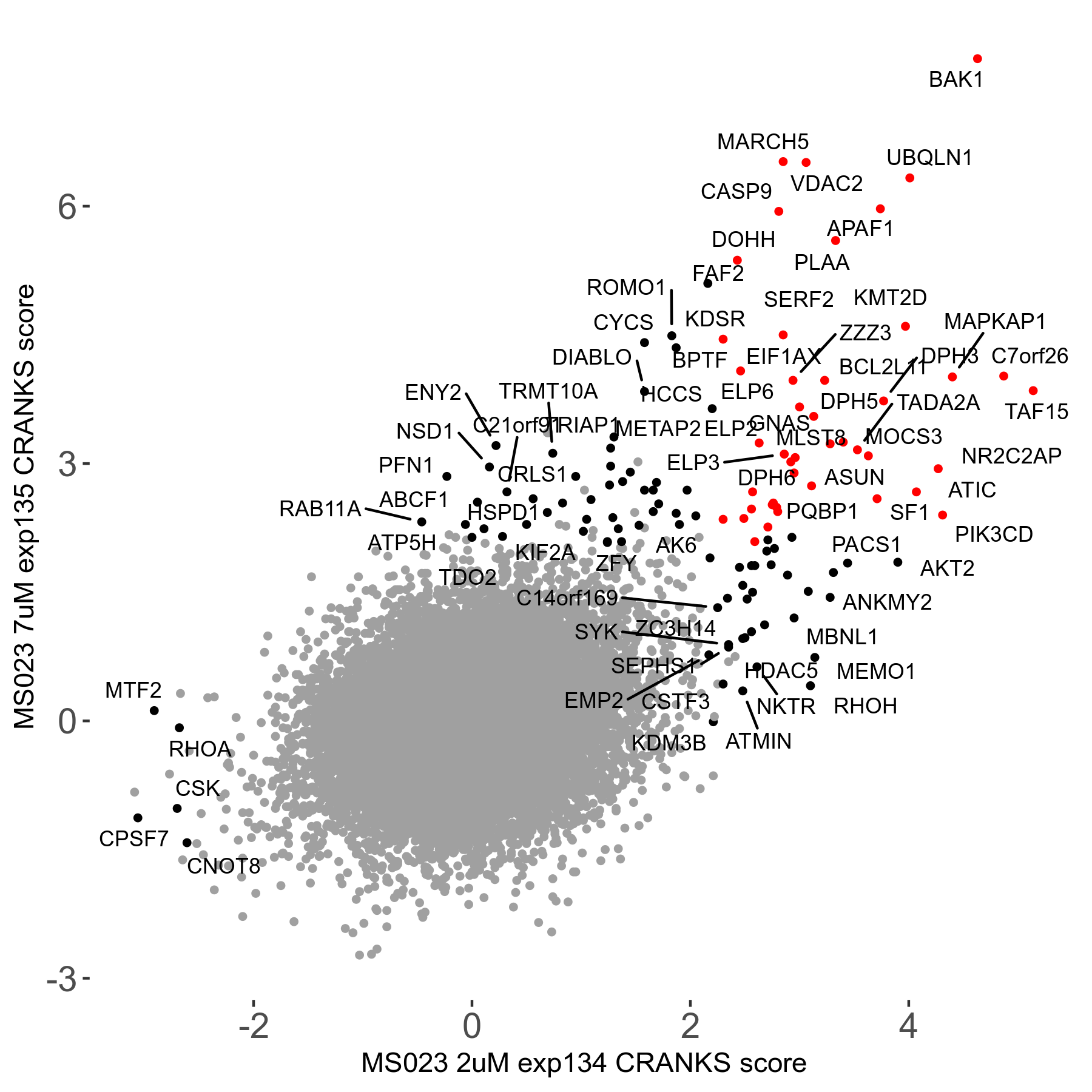
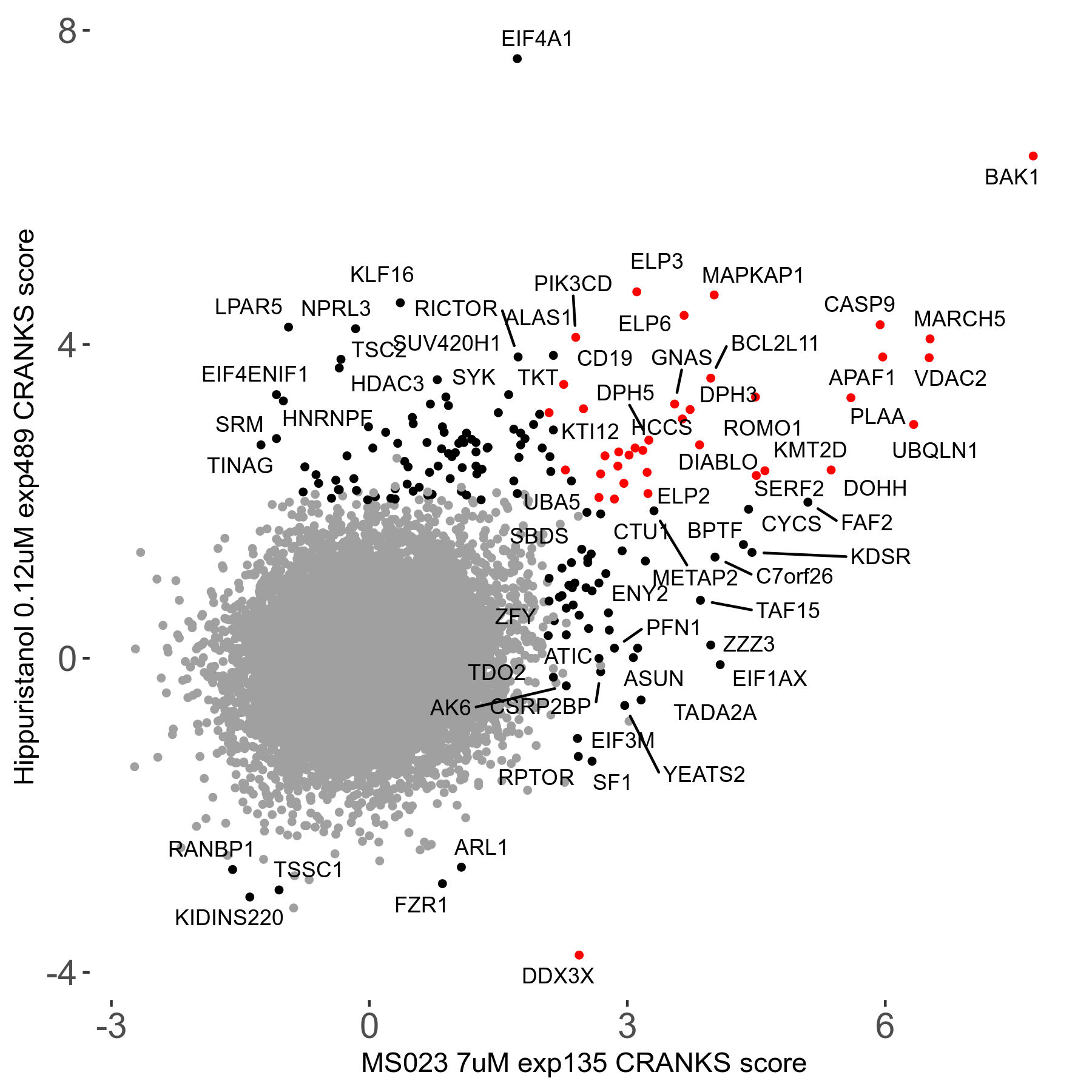
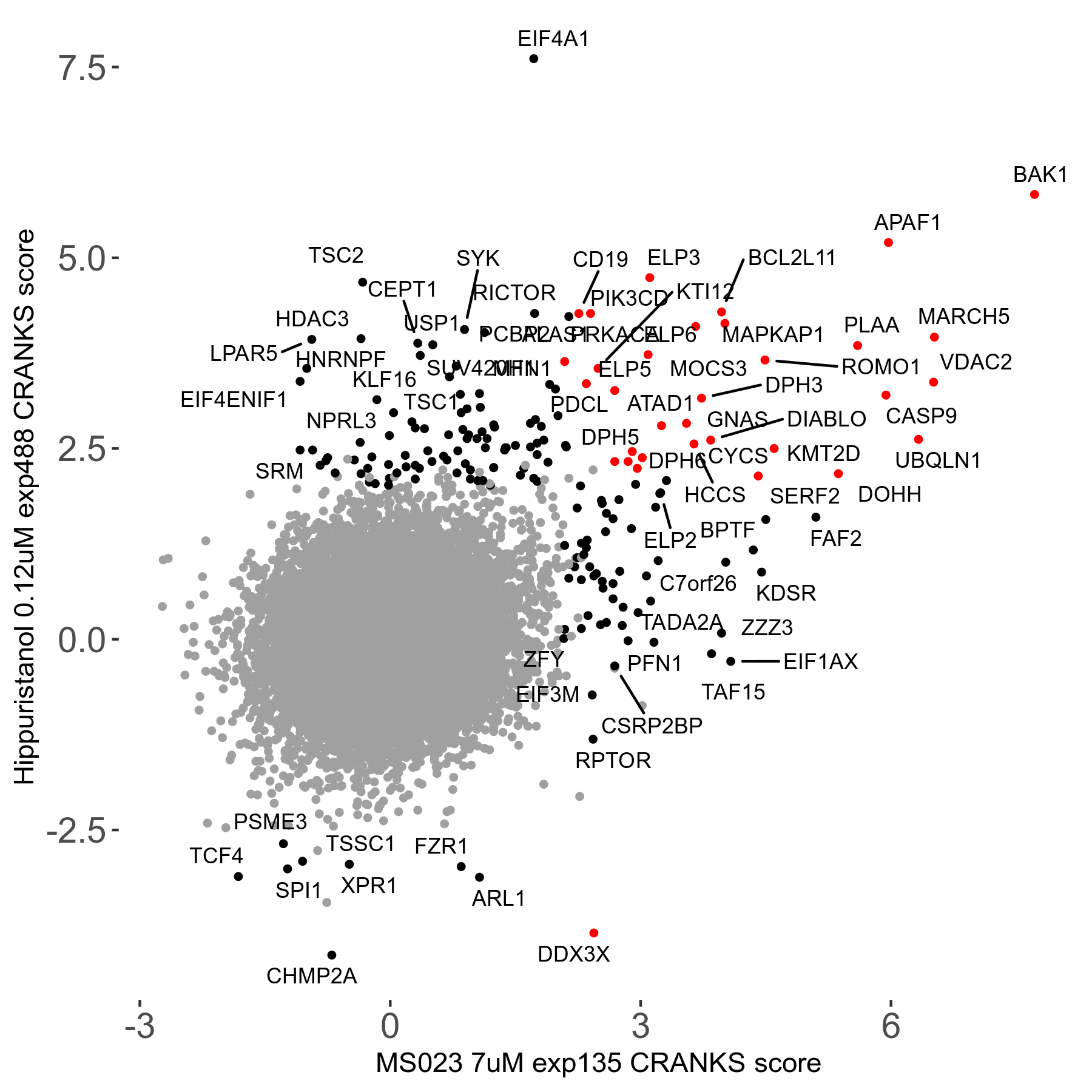
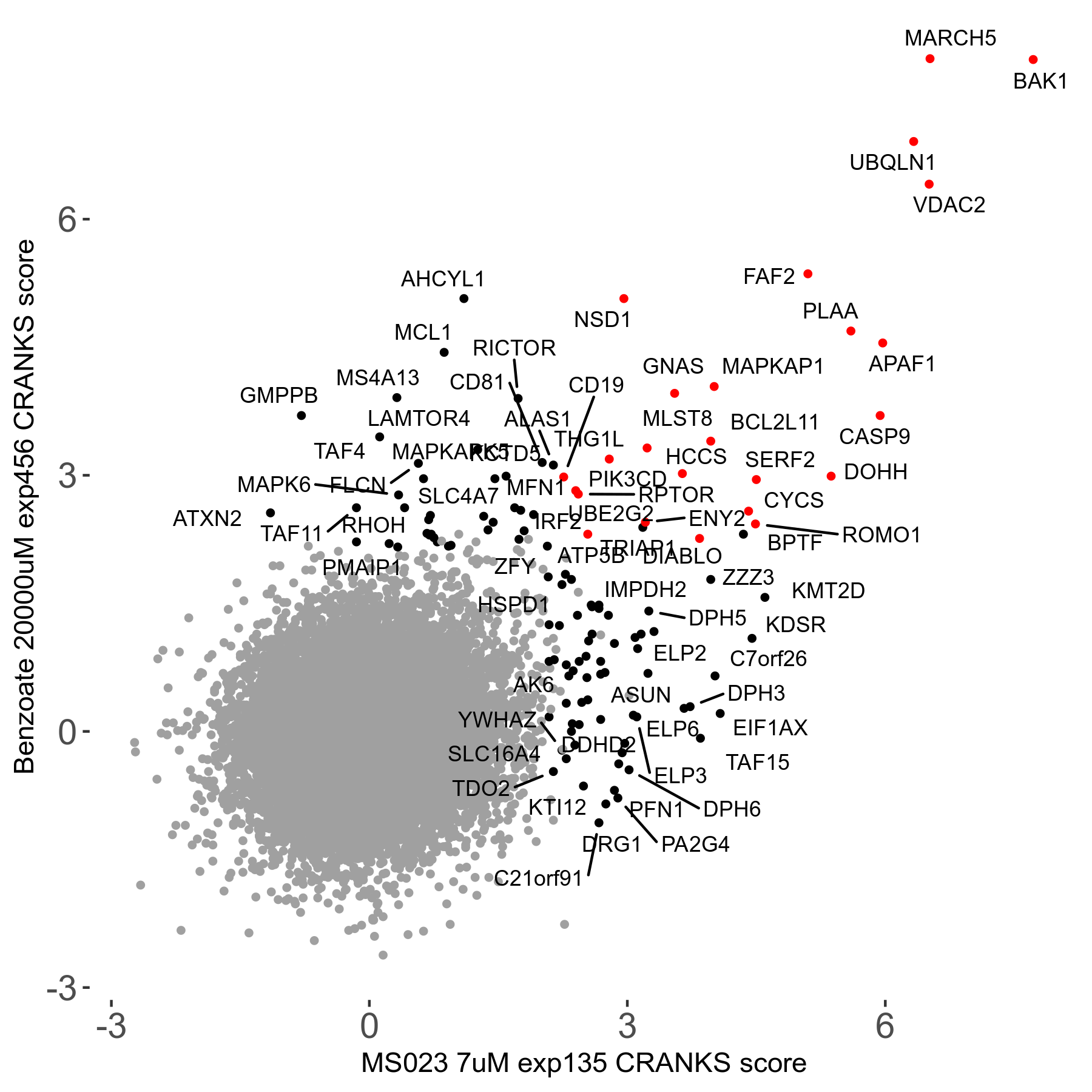
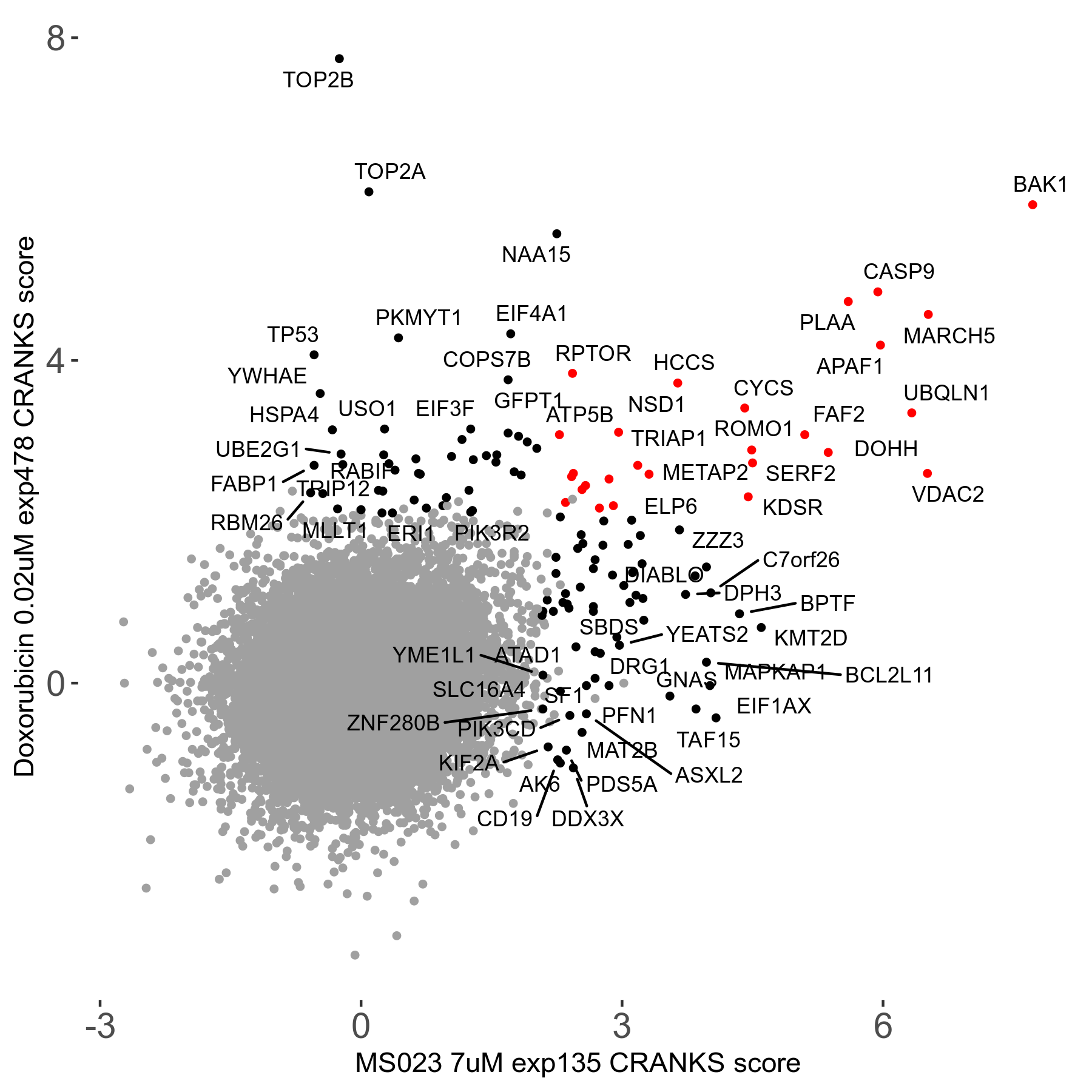
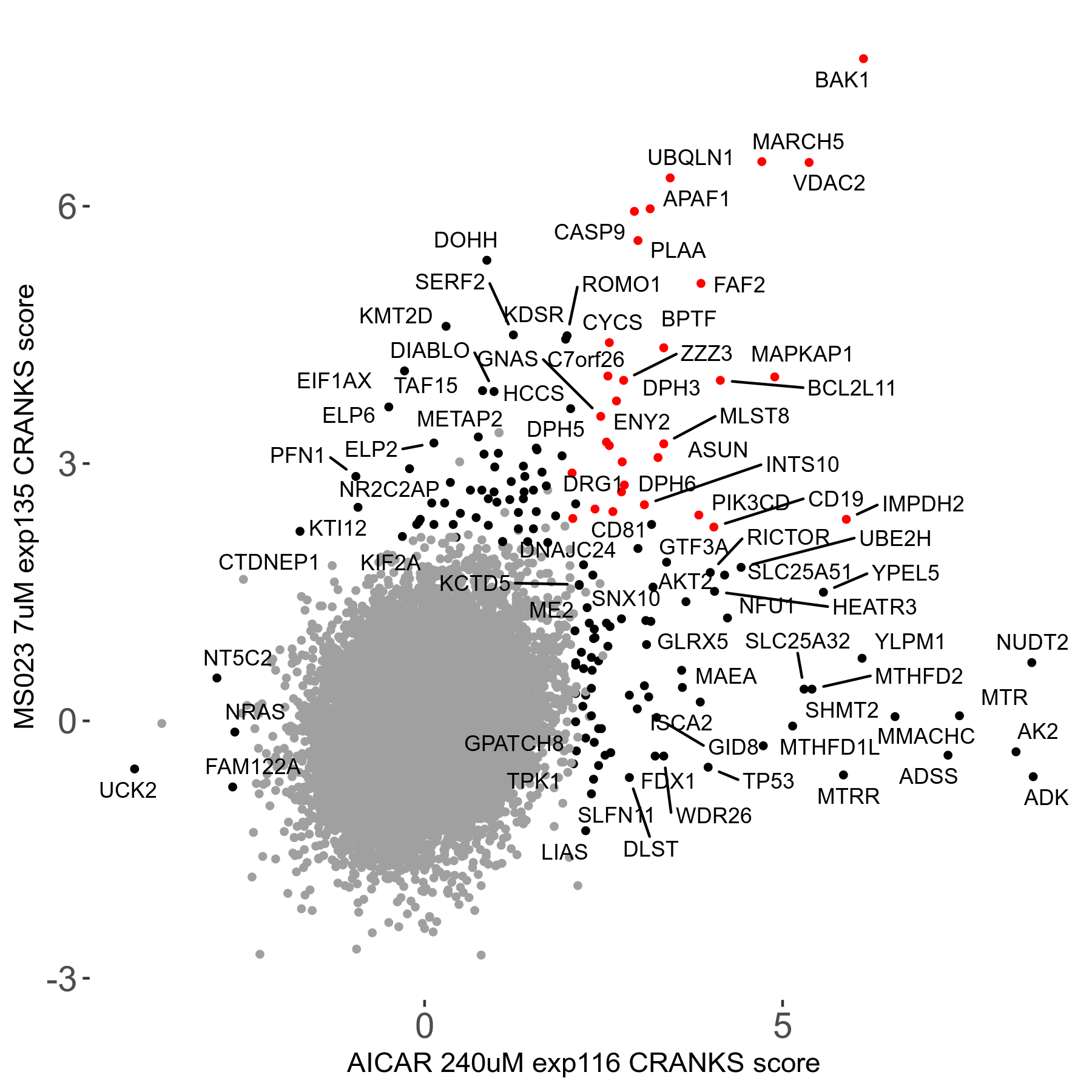
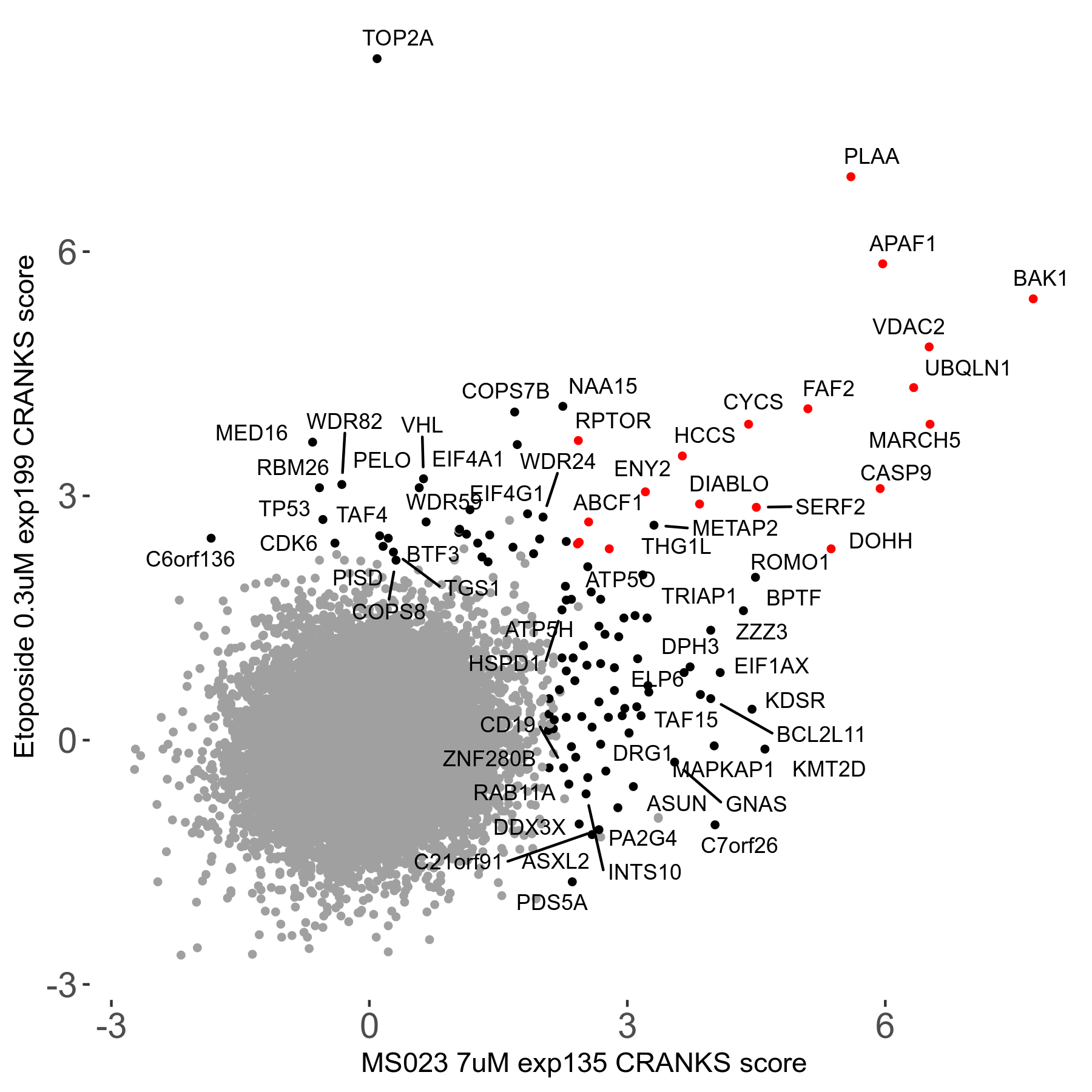
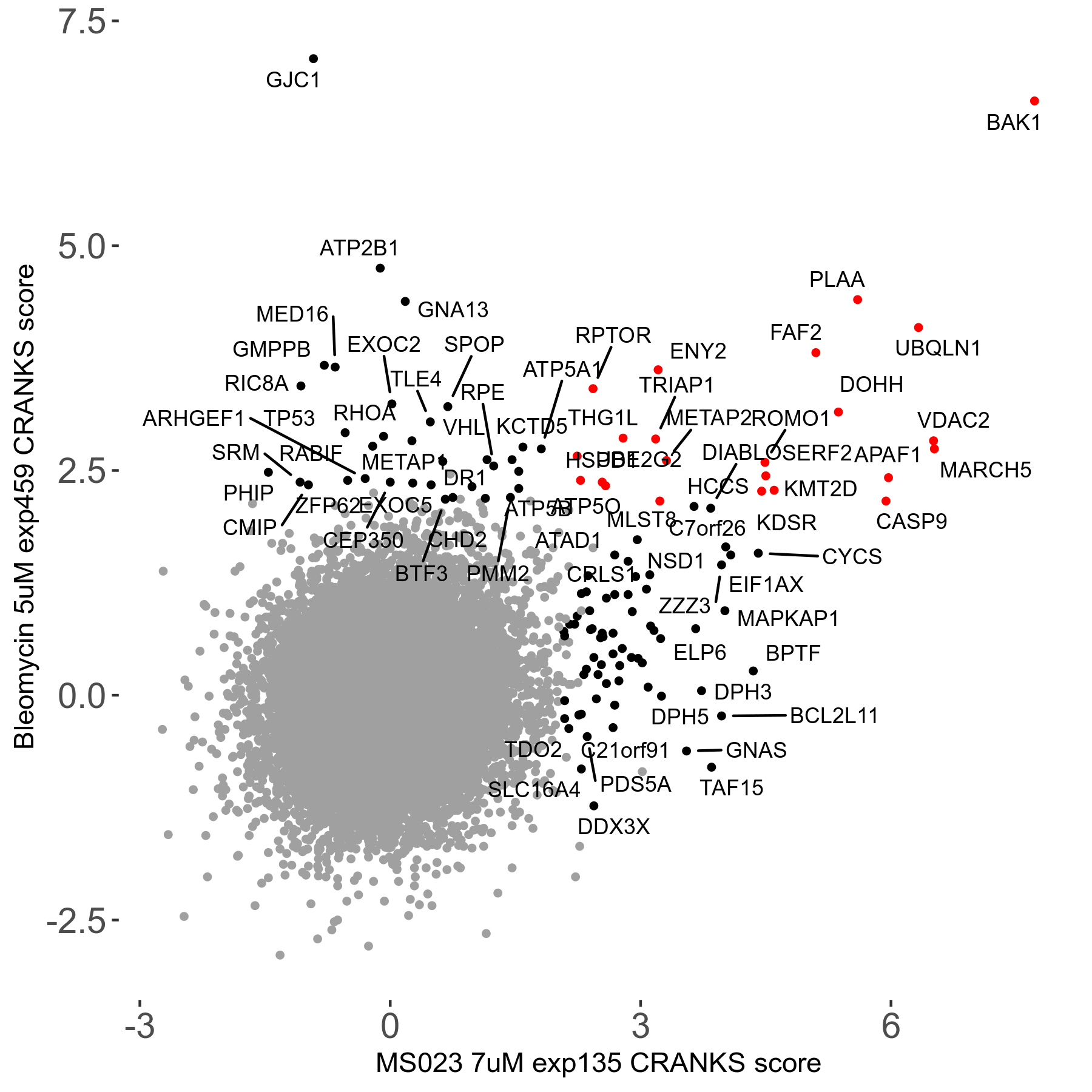
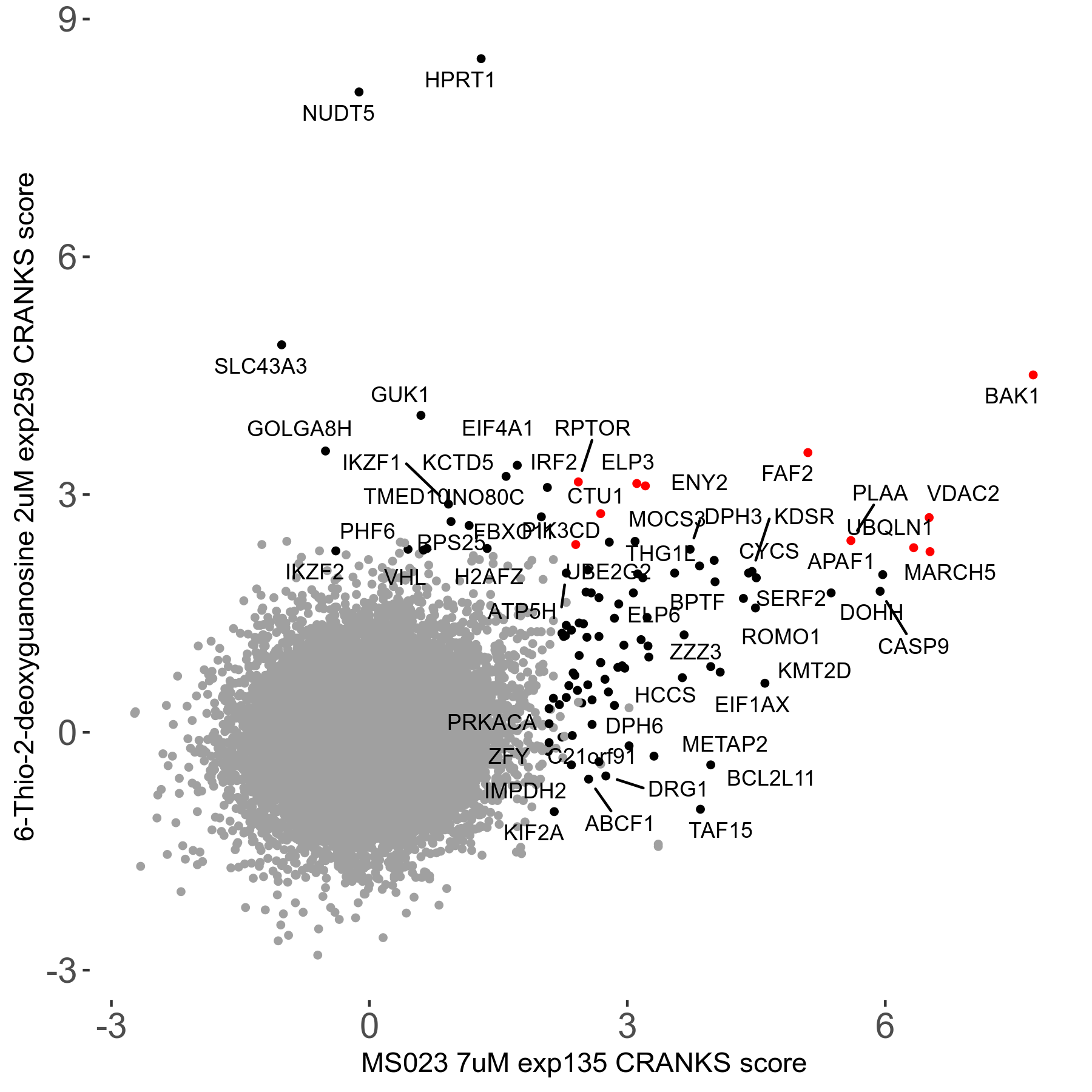
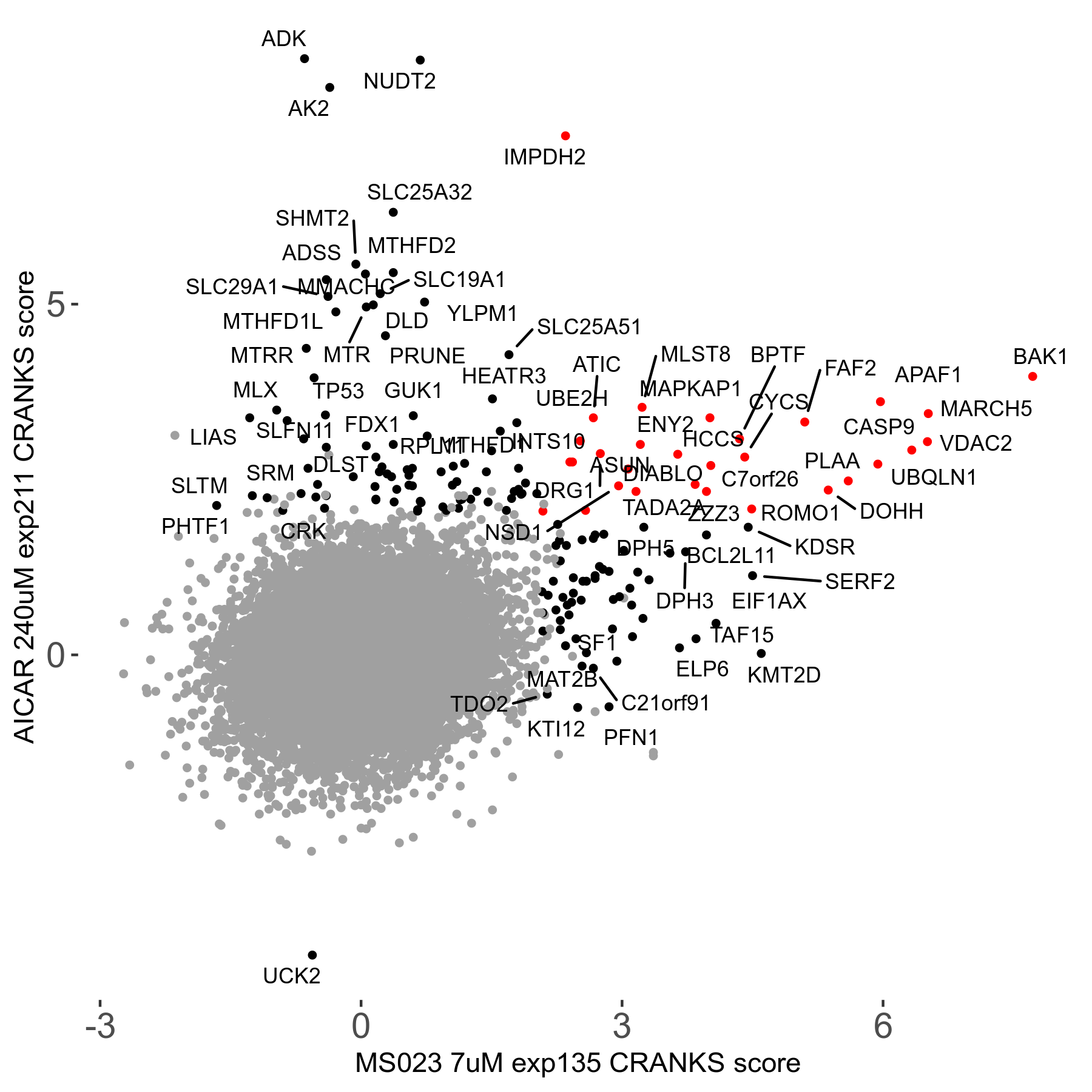
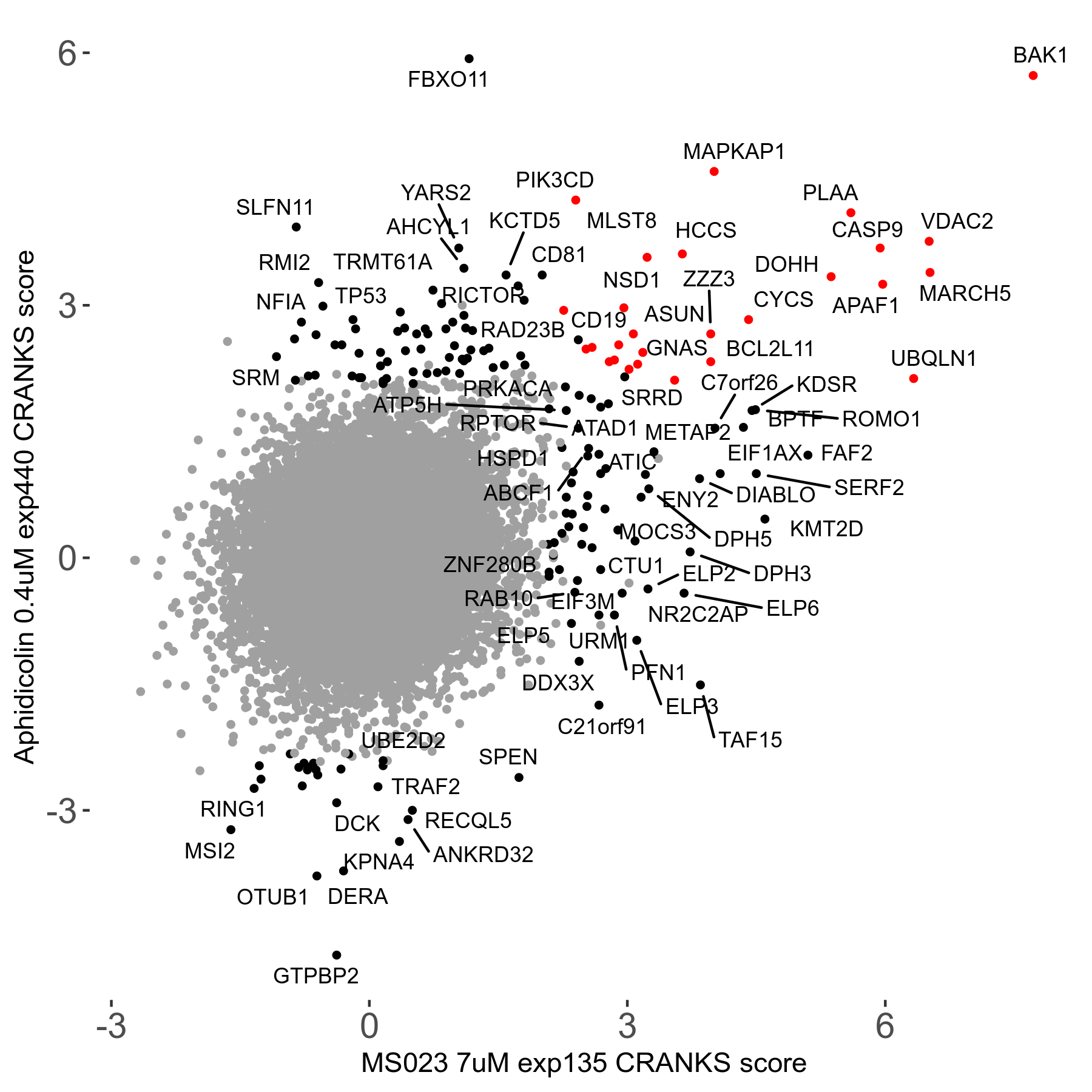
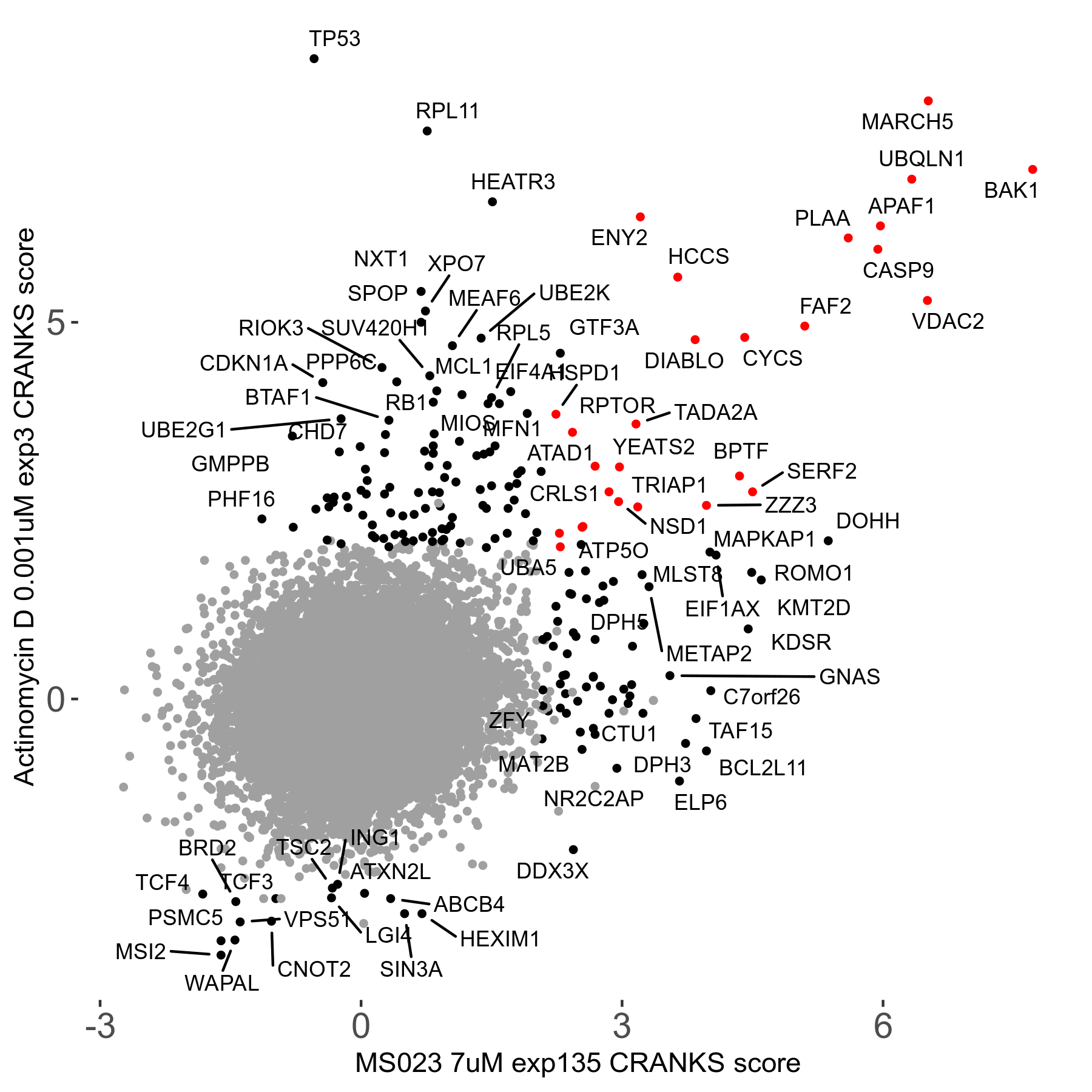
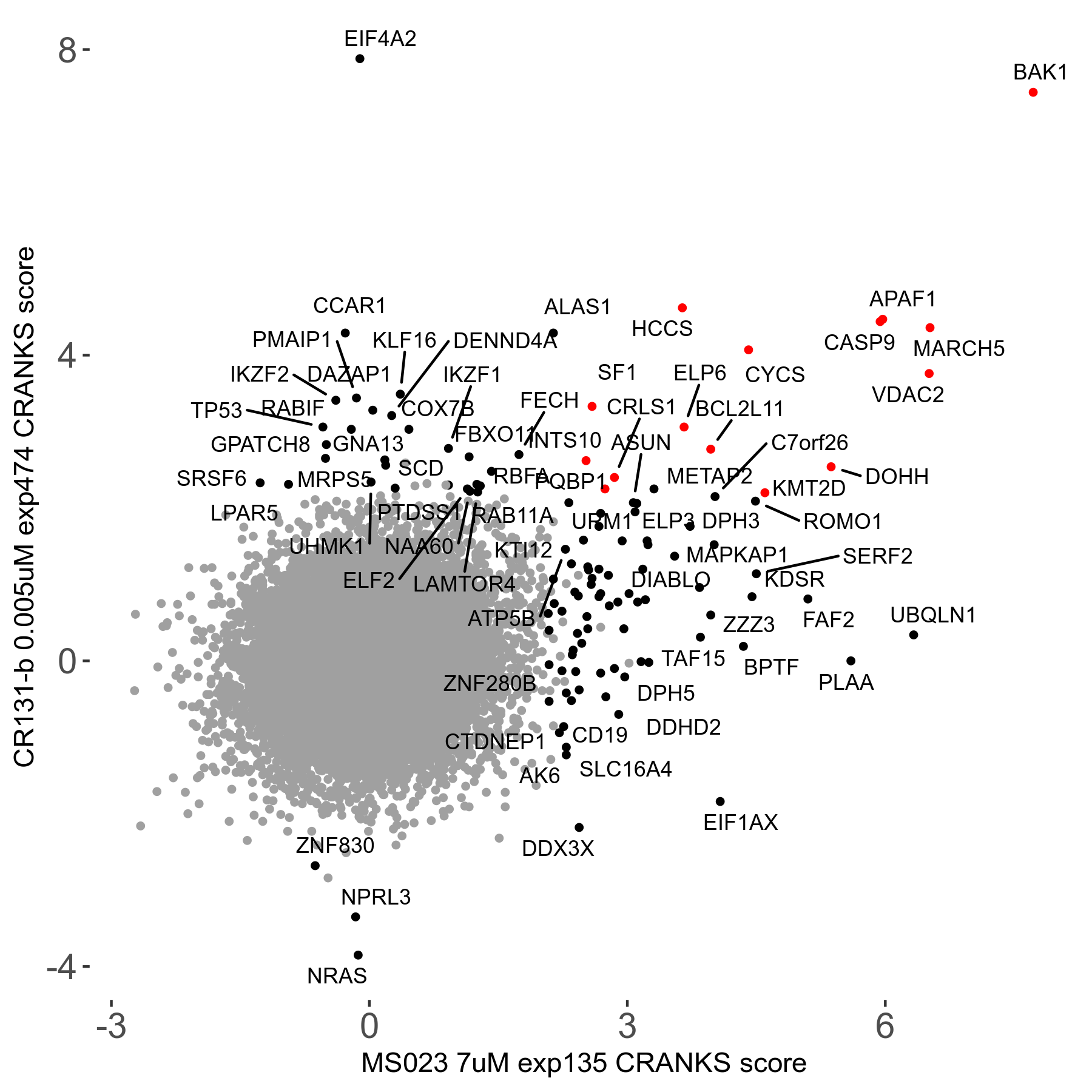
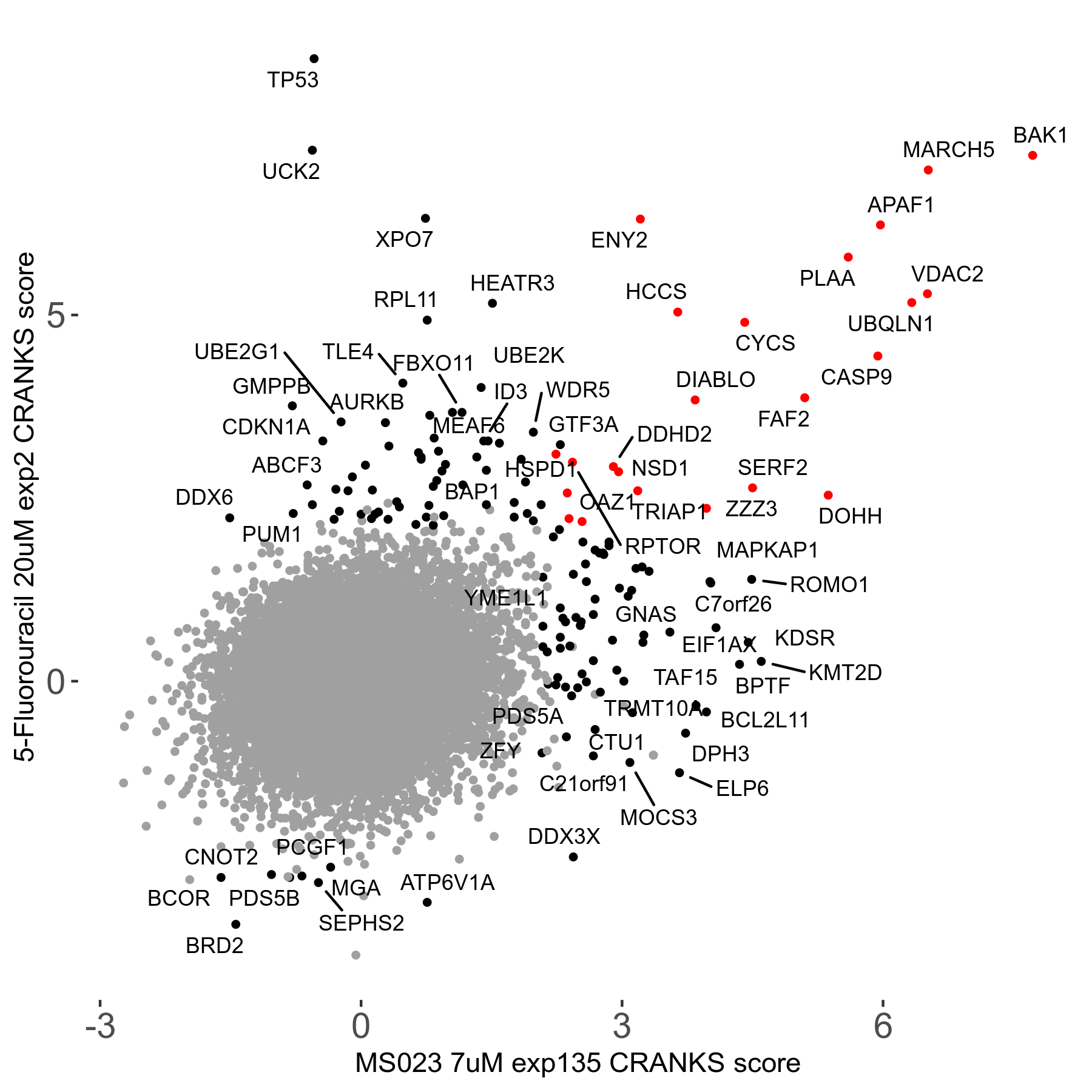
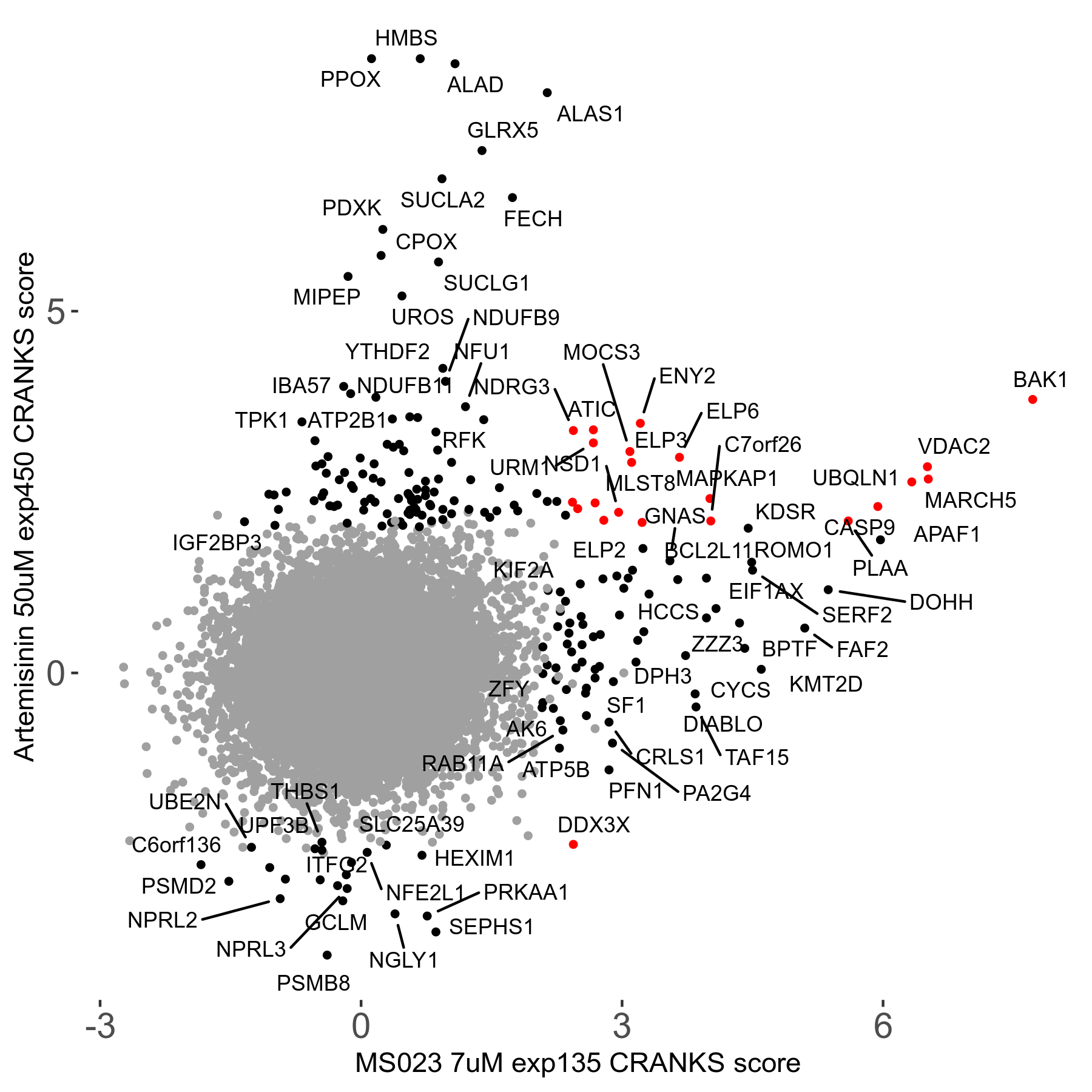
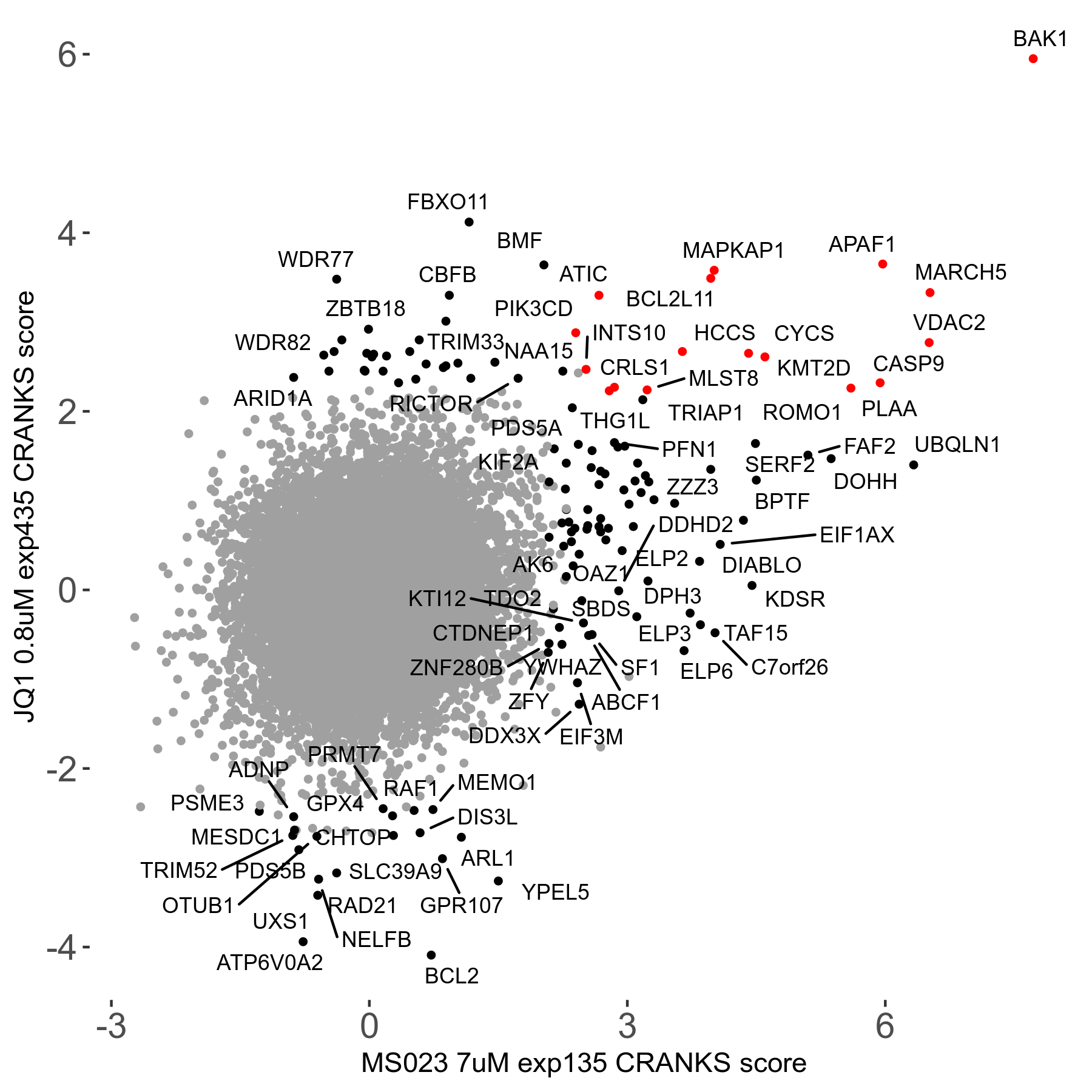
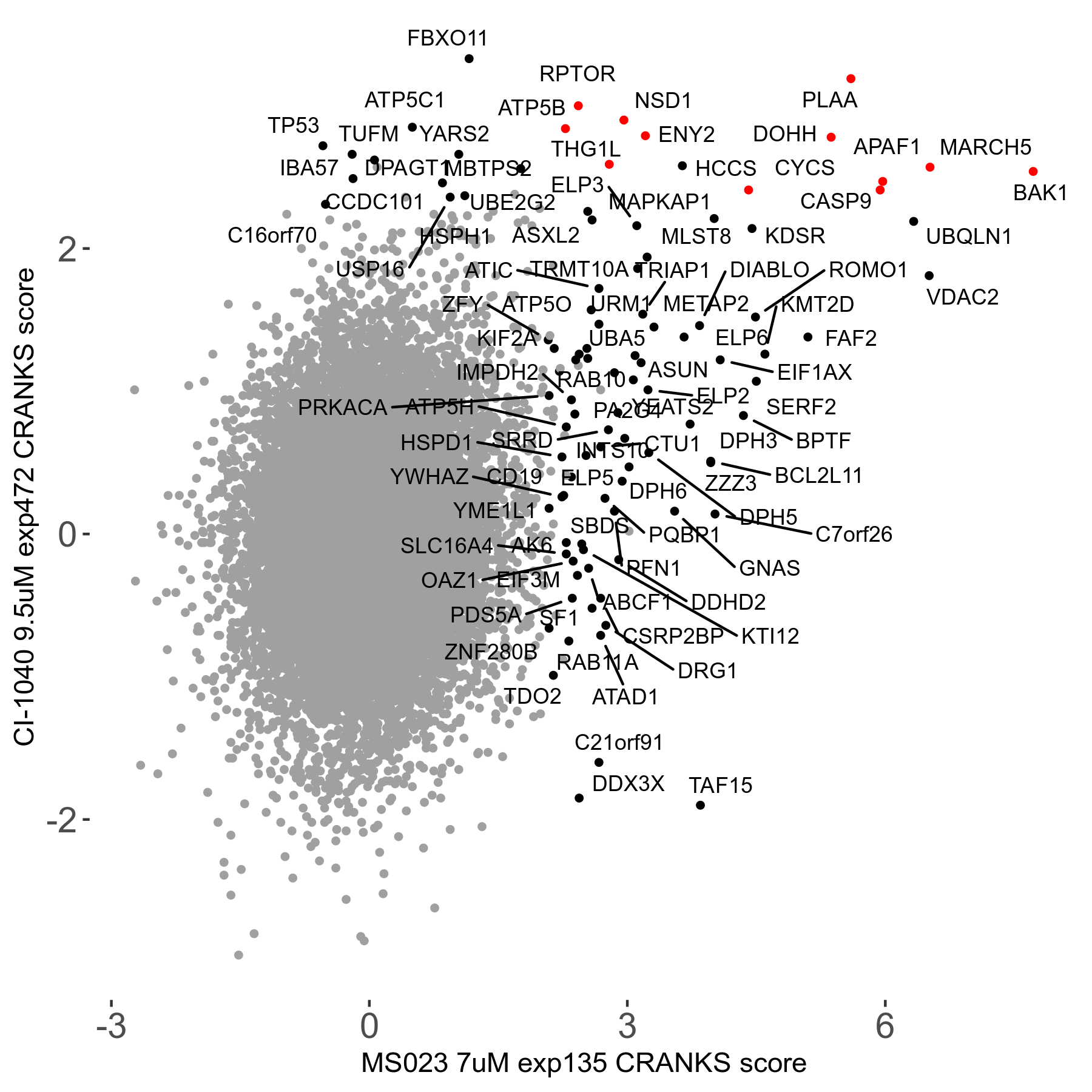
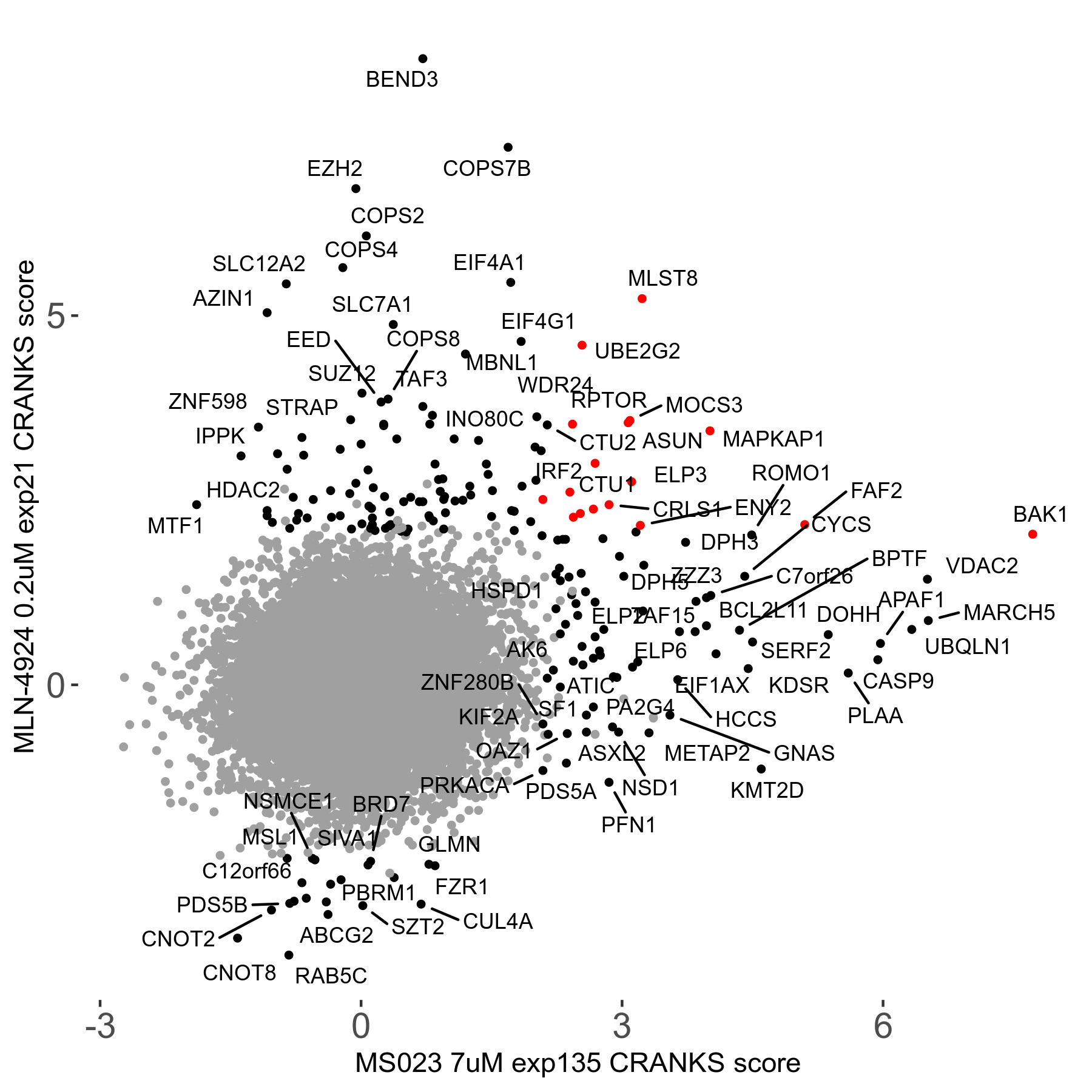
MS023 7μM R03 exp135
Mechanism of Action
Inhibits protein arginine N-methyltransferase PRMT1
- Class / Subclass 1: Gene Regulation / Epigenetic Inhibitor
Technical Notes
Compound References
- PubChem Name: MS023 (dihydrochloride)
- Synonyms: N/A
- CAS #: 1992047-64-9
- PubChem CID: 121513886
- IUPAC: N'-methyl-N'-[[4-(4-propan-2-yloxyphenyl)-1H-pyrrol-3-yl]methyl]ethane-1,2-diamine;dihydrochloride
- INCHI Name: InChI=1S/C17H25N3O.2ClH/c1-13(2)21-16-6-4-14(5-7-16)17-11-19-10-15(17)12-20(3)9-8-18;;/h4-7,10-11,13,19H,8-9,12,18H2,1-3H3;2*1H
- INCHI Key: HCNXCUFNZWGILO-UHFFFAOYSA-N
- Molecular Weight: 360.3
- Canonical SMILES: CC(C)OC1=CC=C(C=C1)C2=CNC=C2CN(C)CCN.Cl.Cl
- Isomeric SMILES: N/A
- Molecular Formula: C17H27Cl2N3O
Compound Supplier
- Supplier Name: Structural Genomics Consortium
- Catalog #: N/A
- Lot #: N/A
Compound Characterization
- HRMS (ESI-TOF) m/z: (M+H)+ Calcd for C17H25N3O 288.20704; found 288.20735
Dose Response Curve
- Platform ID: MS023
- Min: -6.0031; Max: 44.7664

| IC | Concentration (µM) |
|---|---|
| IC10 | N/A |
| IC20 | 3.5114 |
| IC30 | 7.8192 |
| IC40 | 15.0721 |
| IC50 | N/A |
| IC60 | N/A |
| IC70 | N/A |
| IC80 | N/A |
| IC90 | N/A |
Screen Summary
- Round: 03
- Dose: 7µM
- Days of incubation: 8
- Doublings: 2.6
- Numbers of reads: 13404640
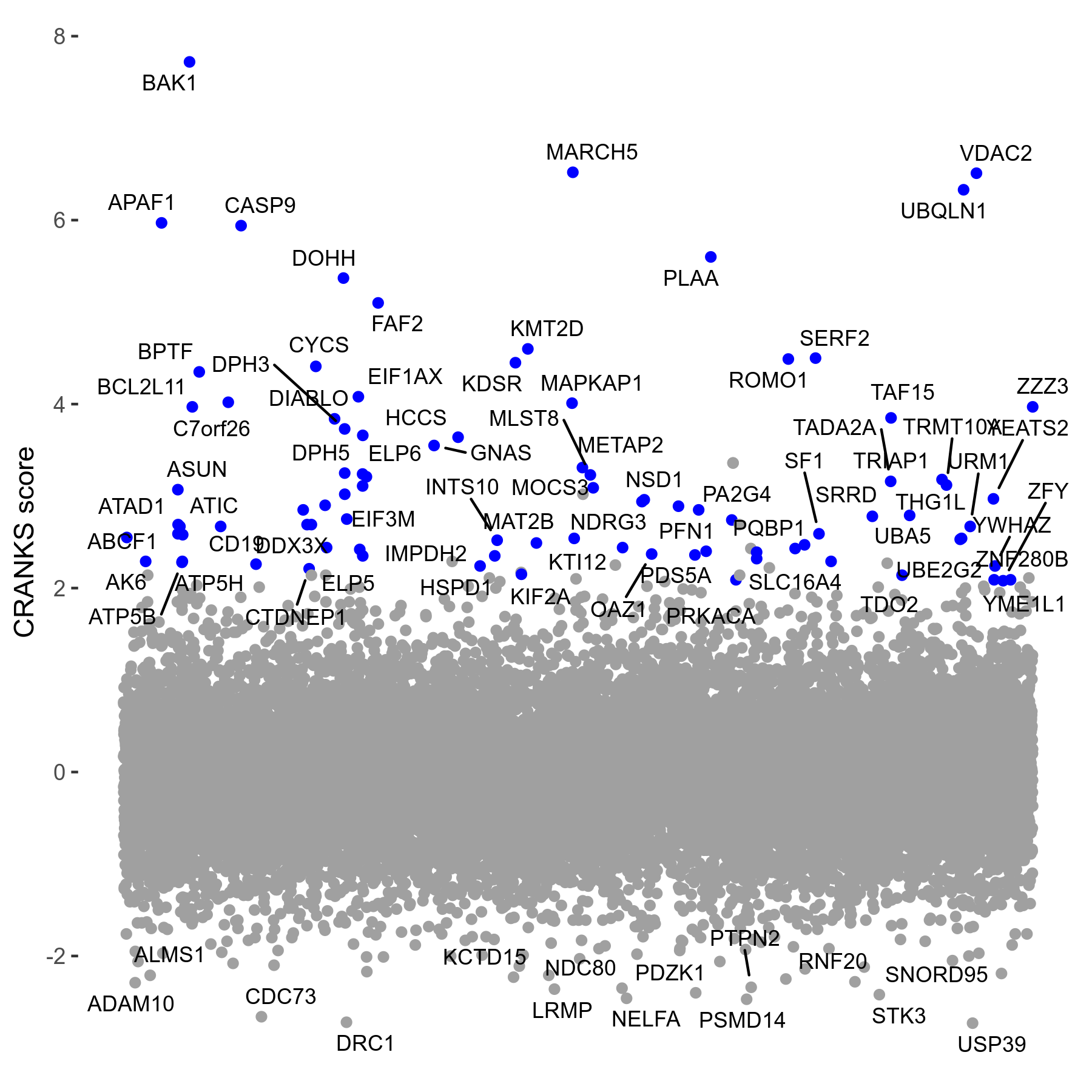
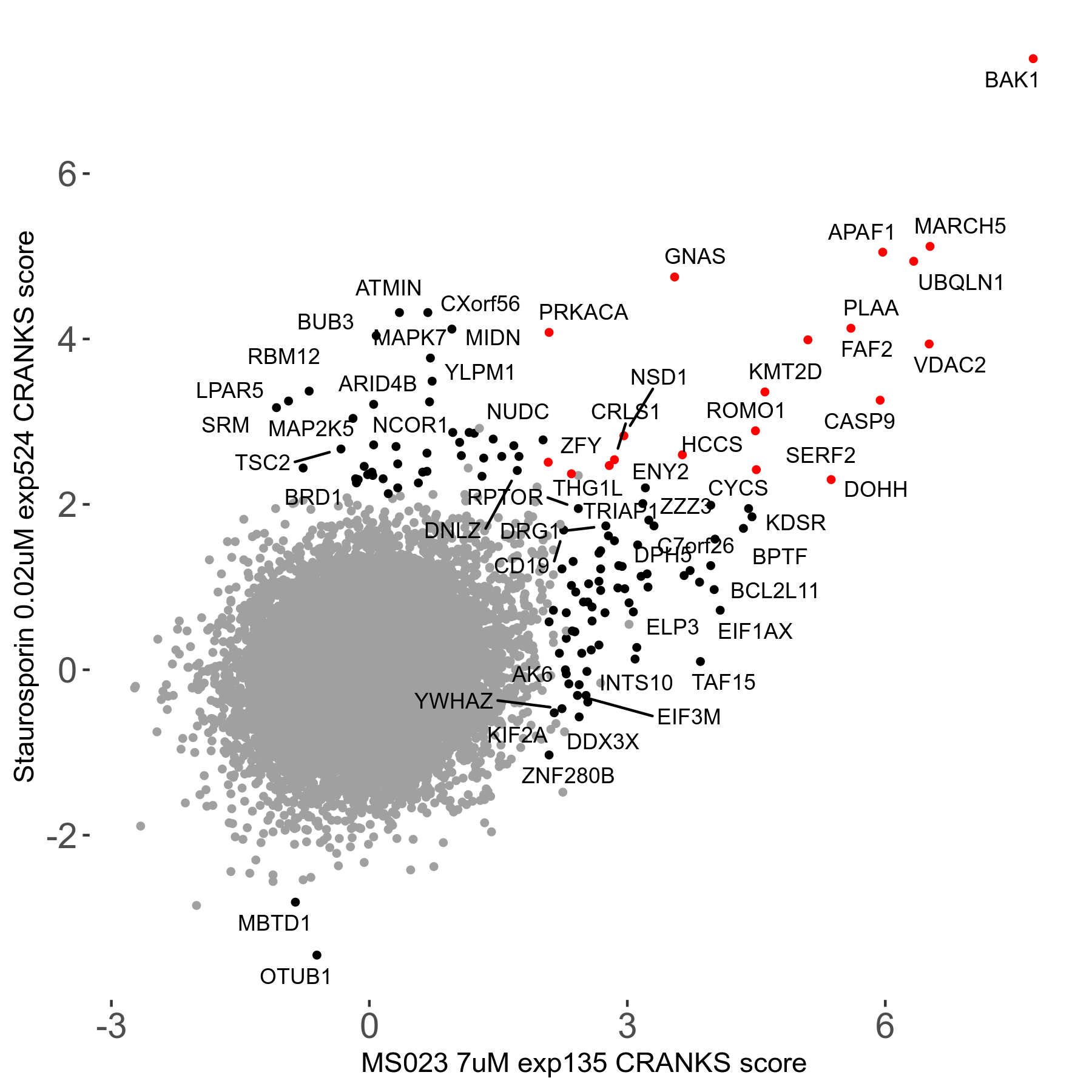
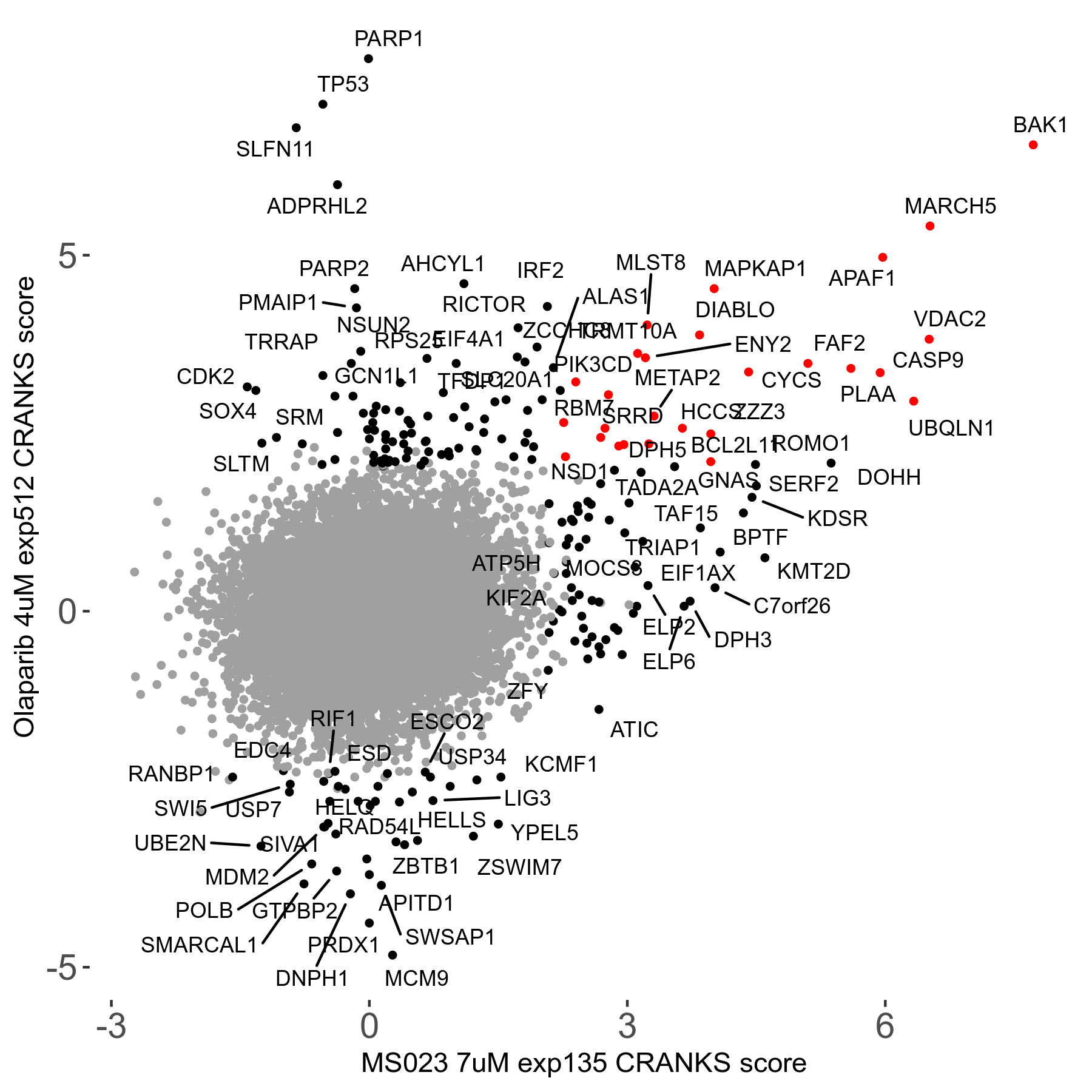
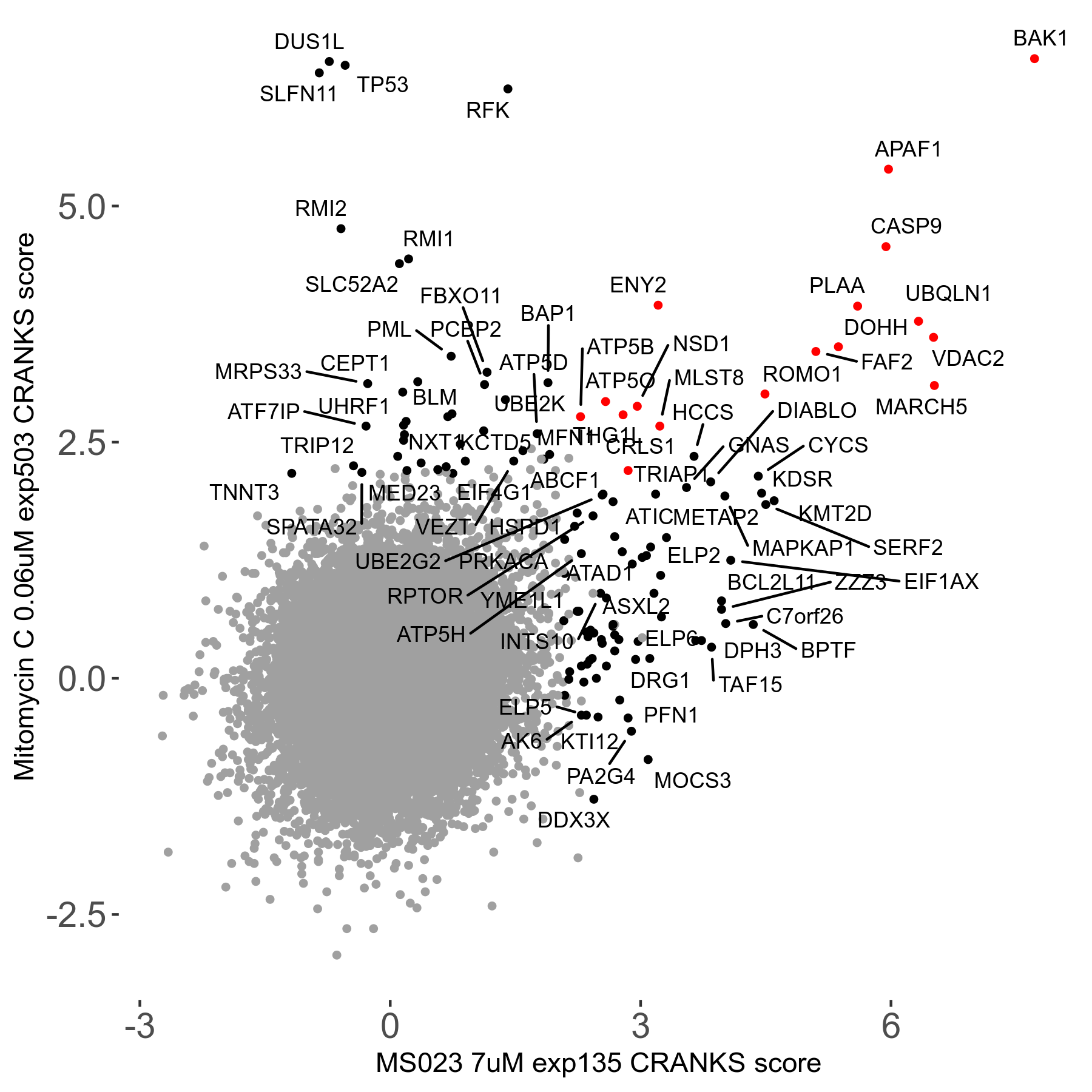
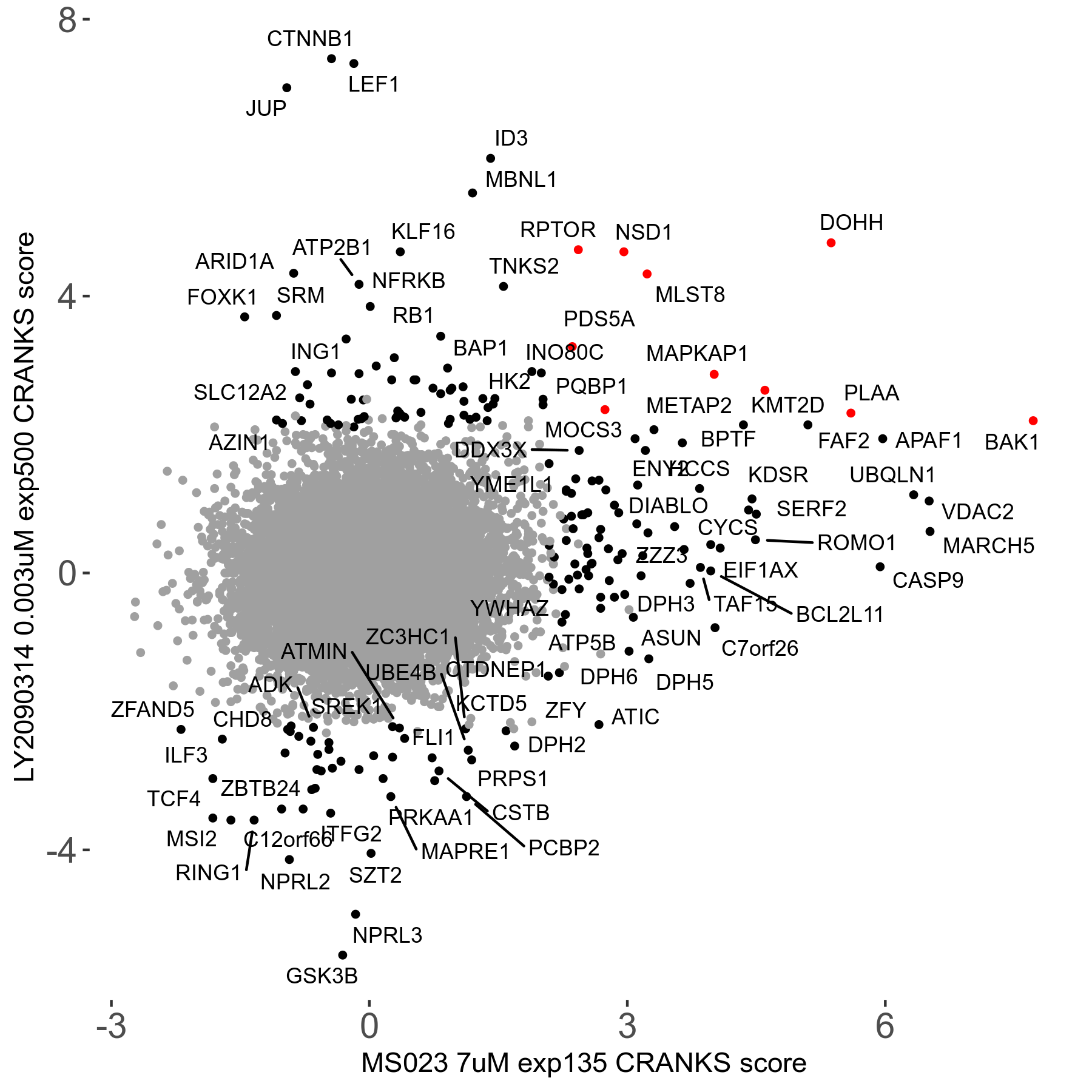
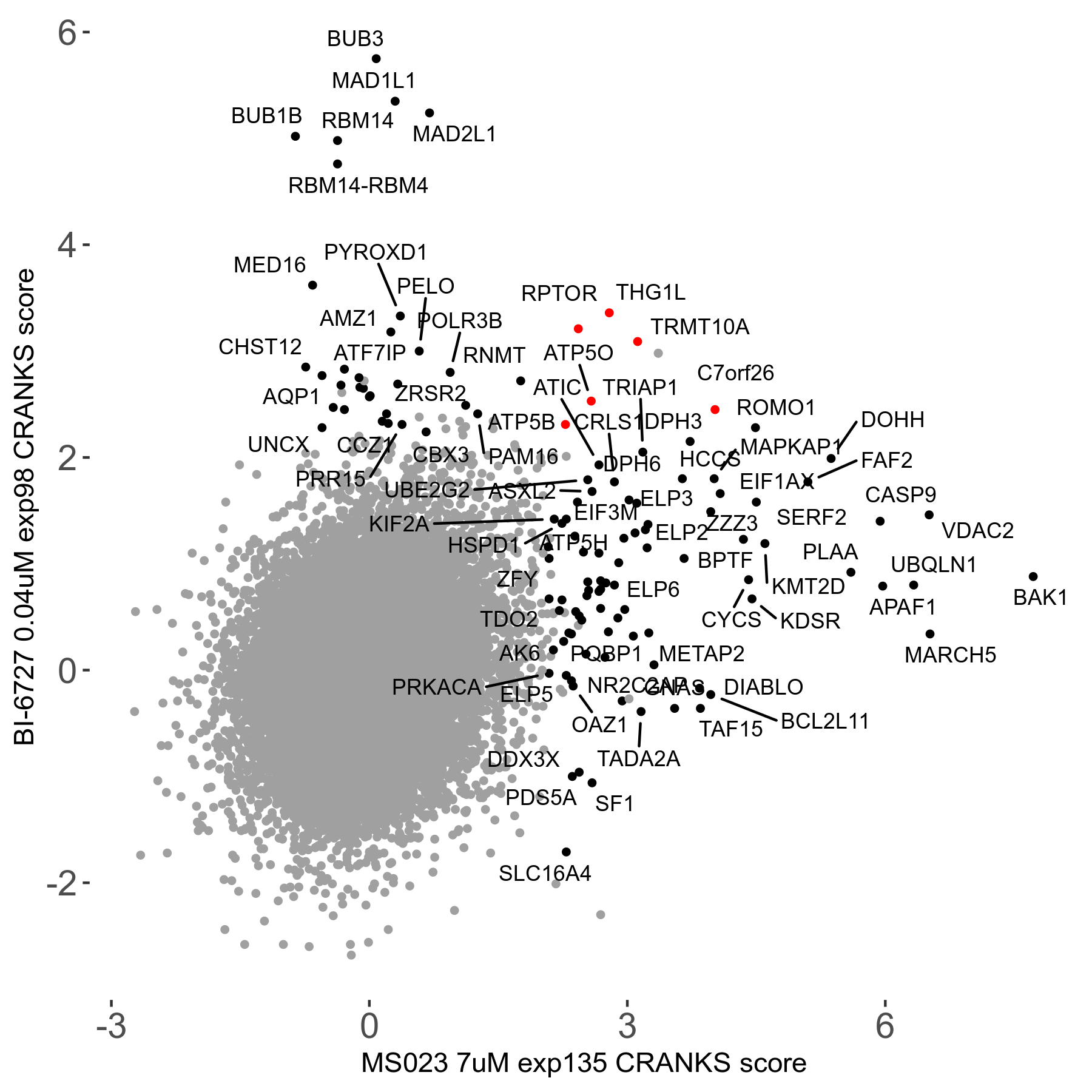
Screen Results
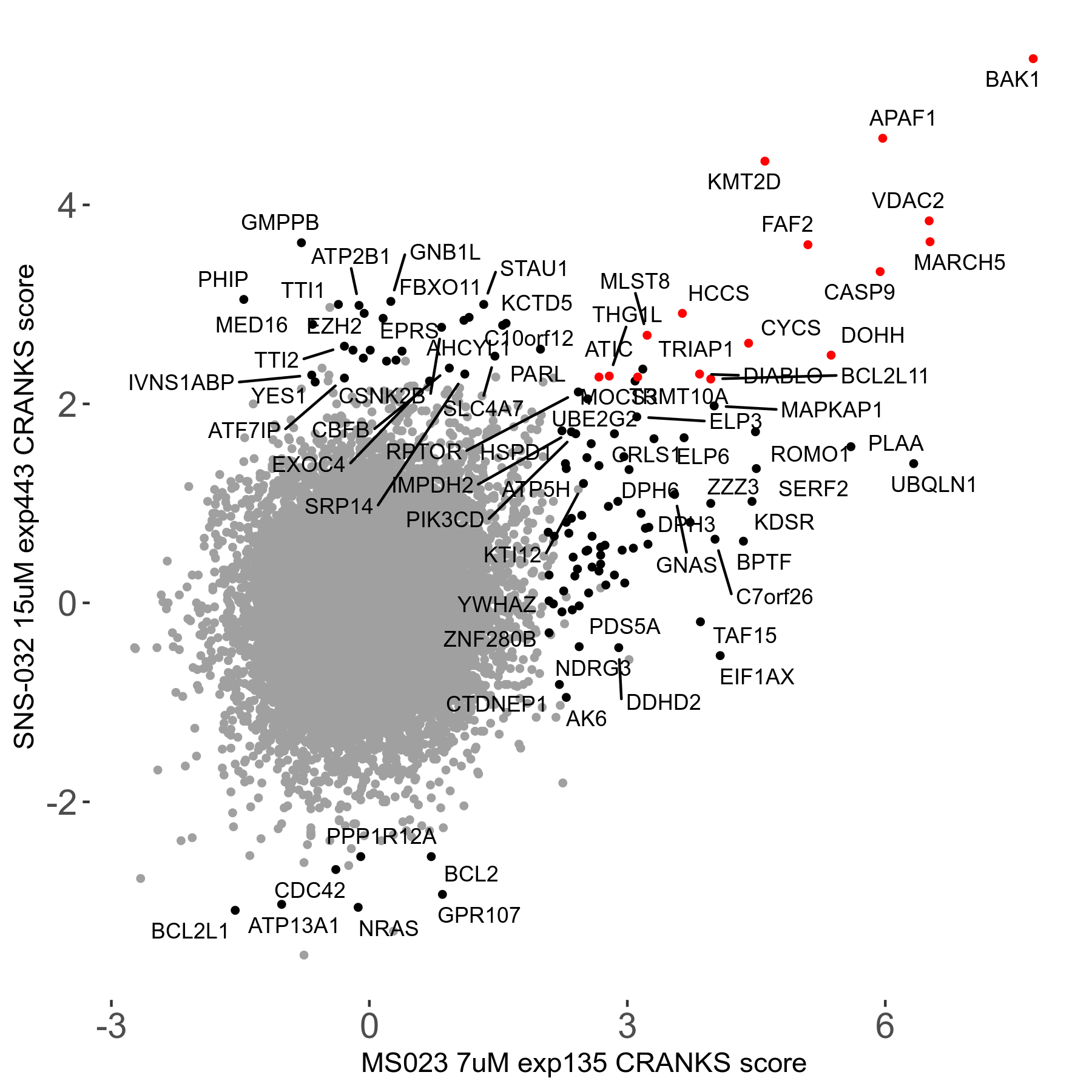
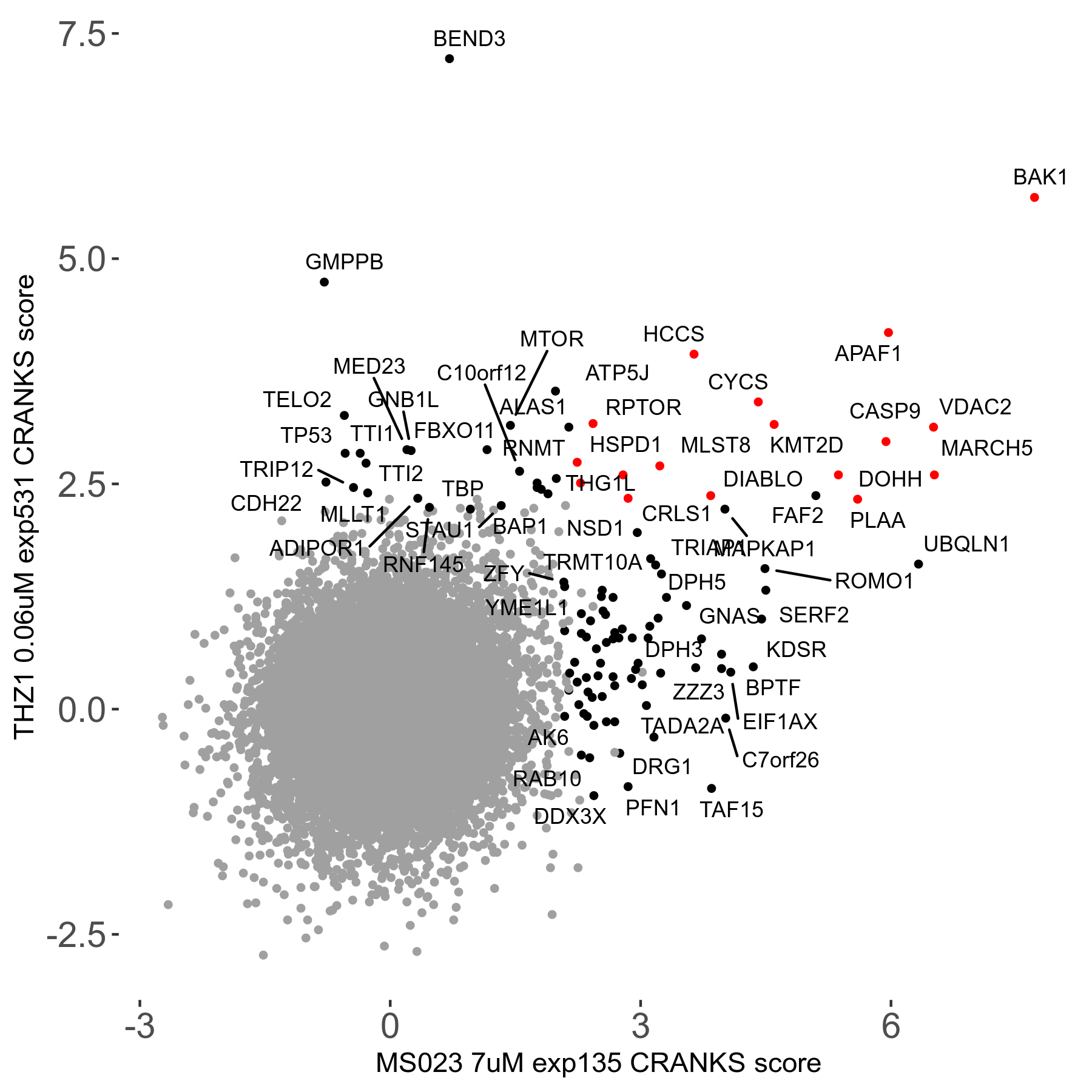
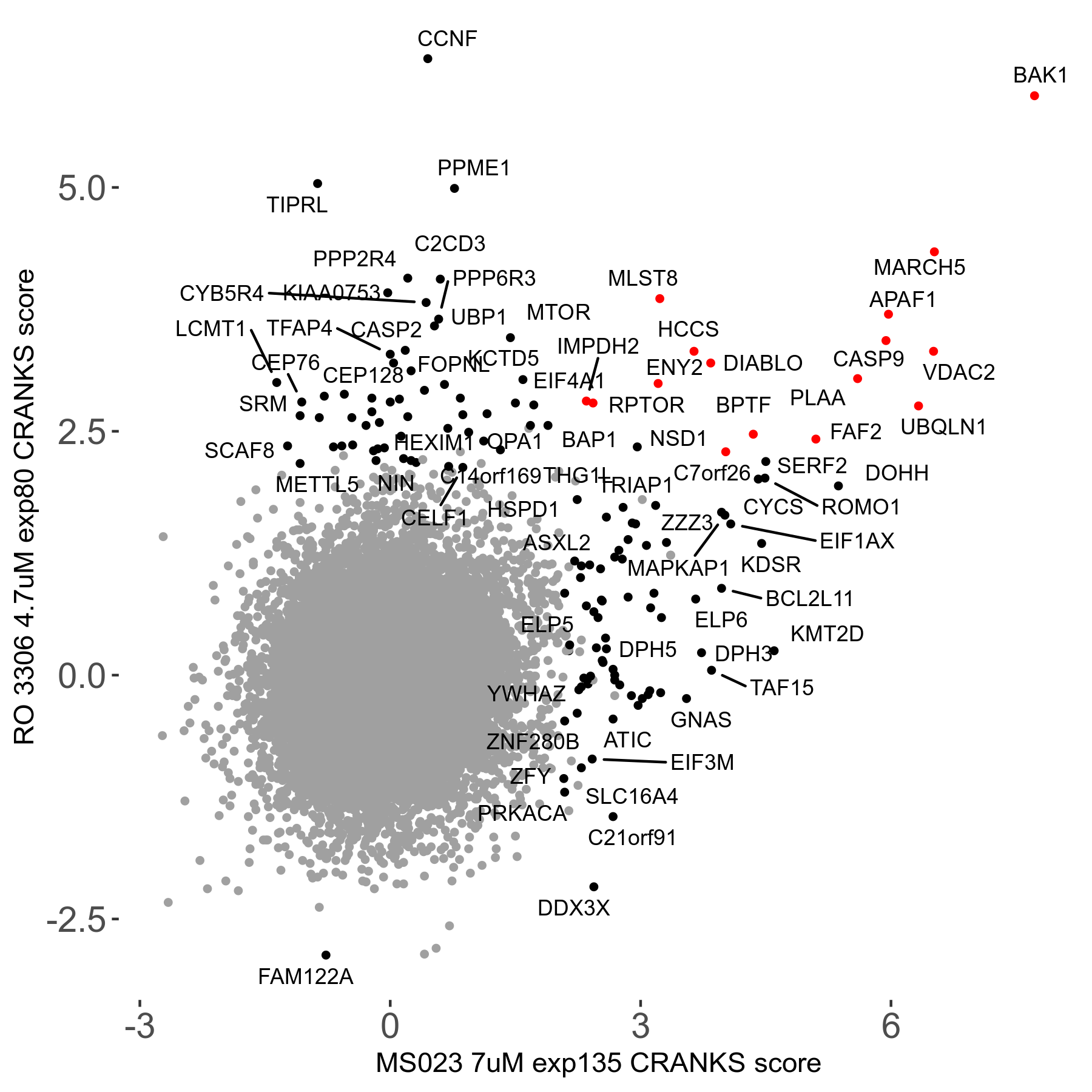
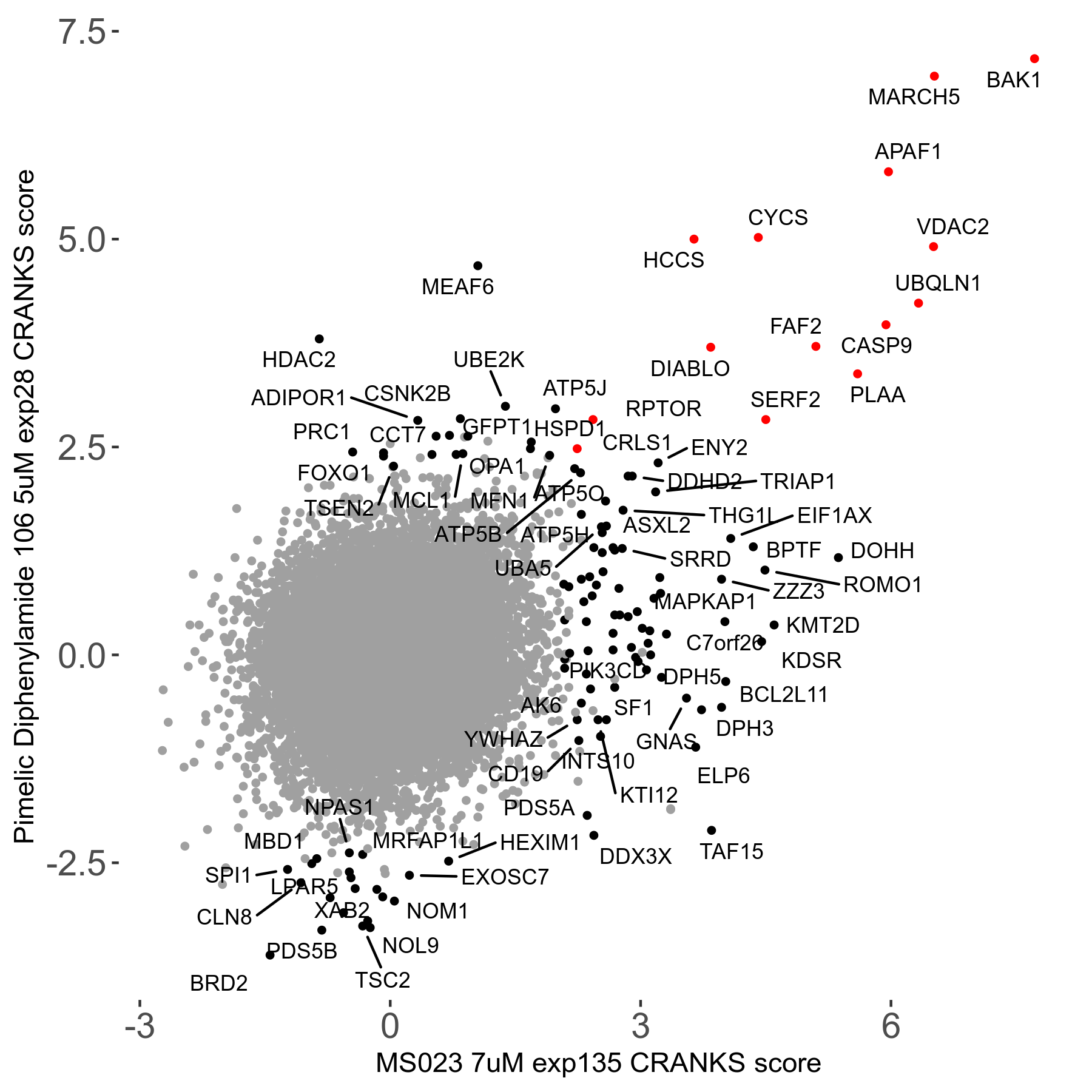
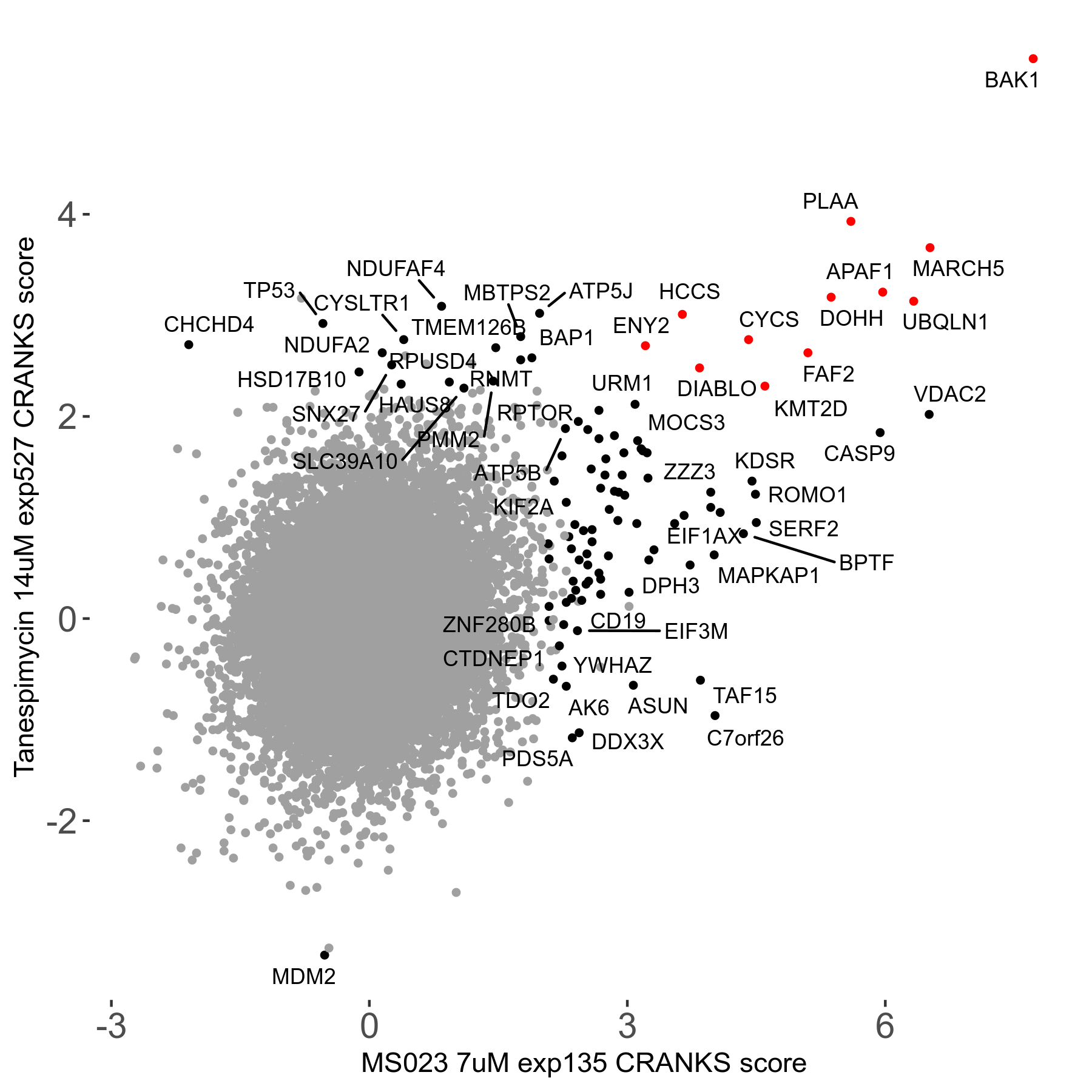
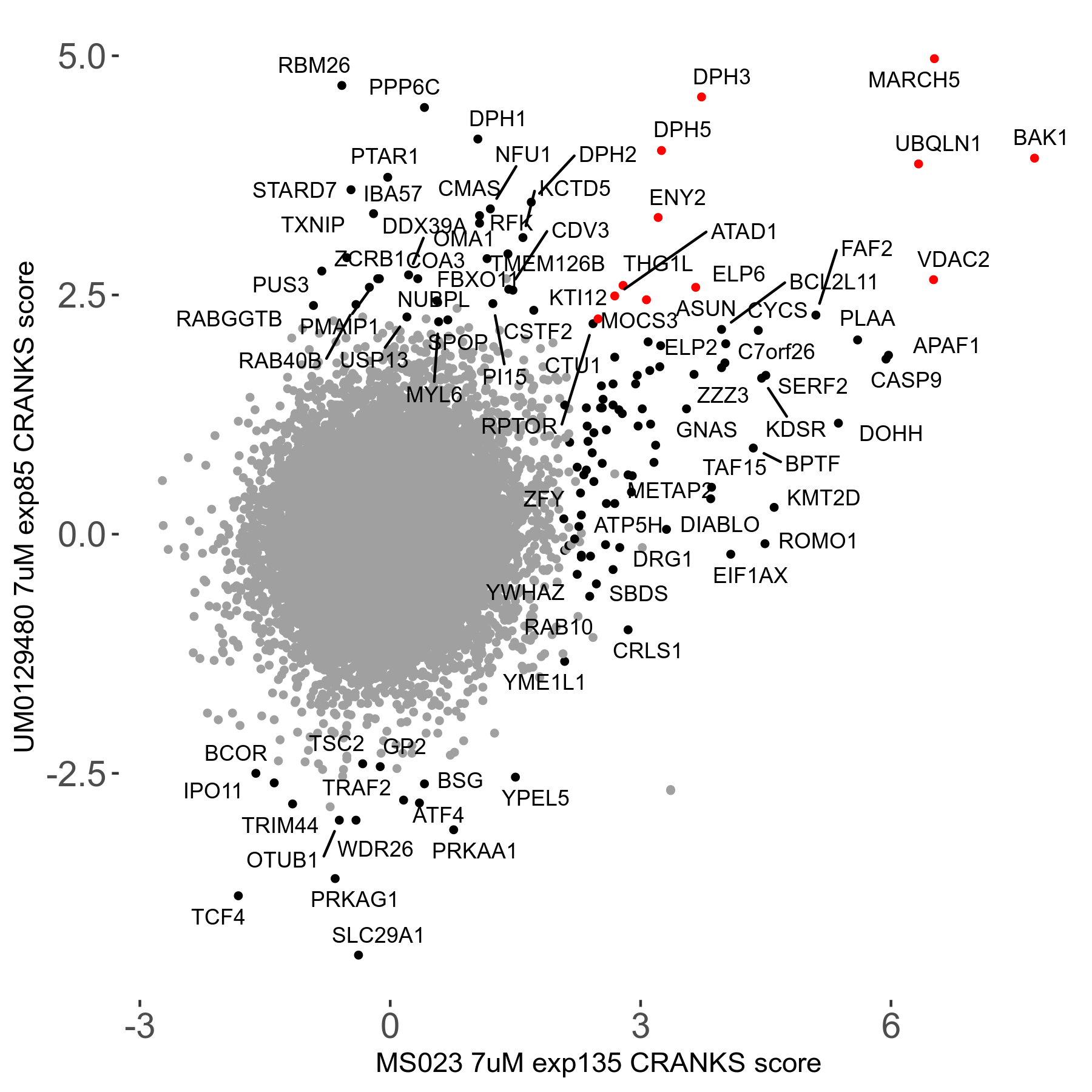
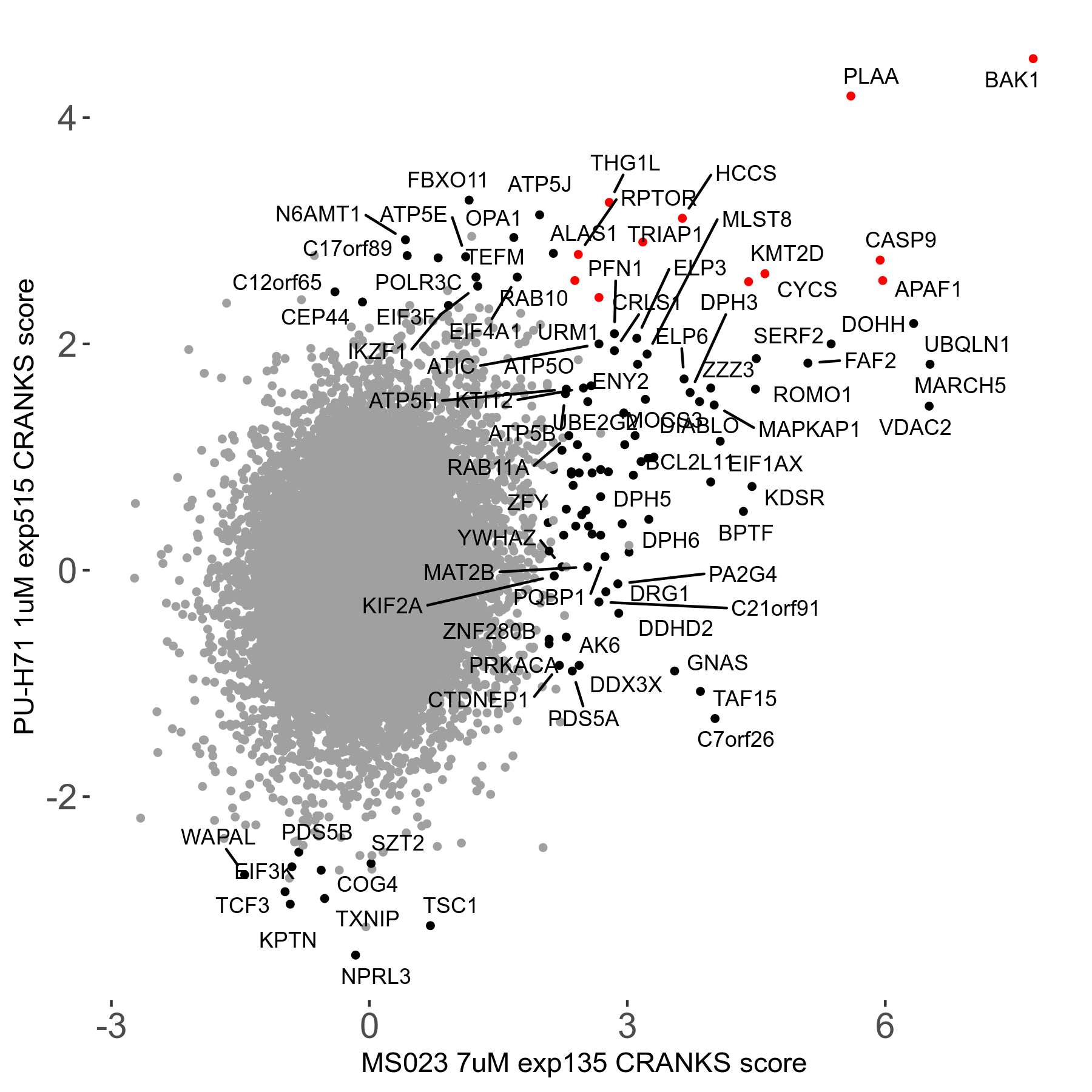
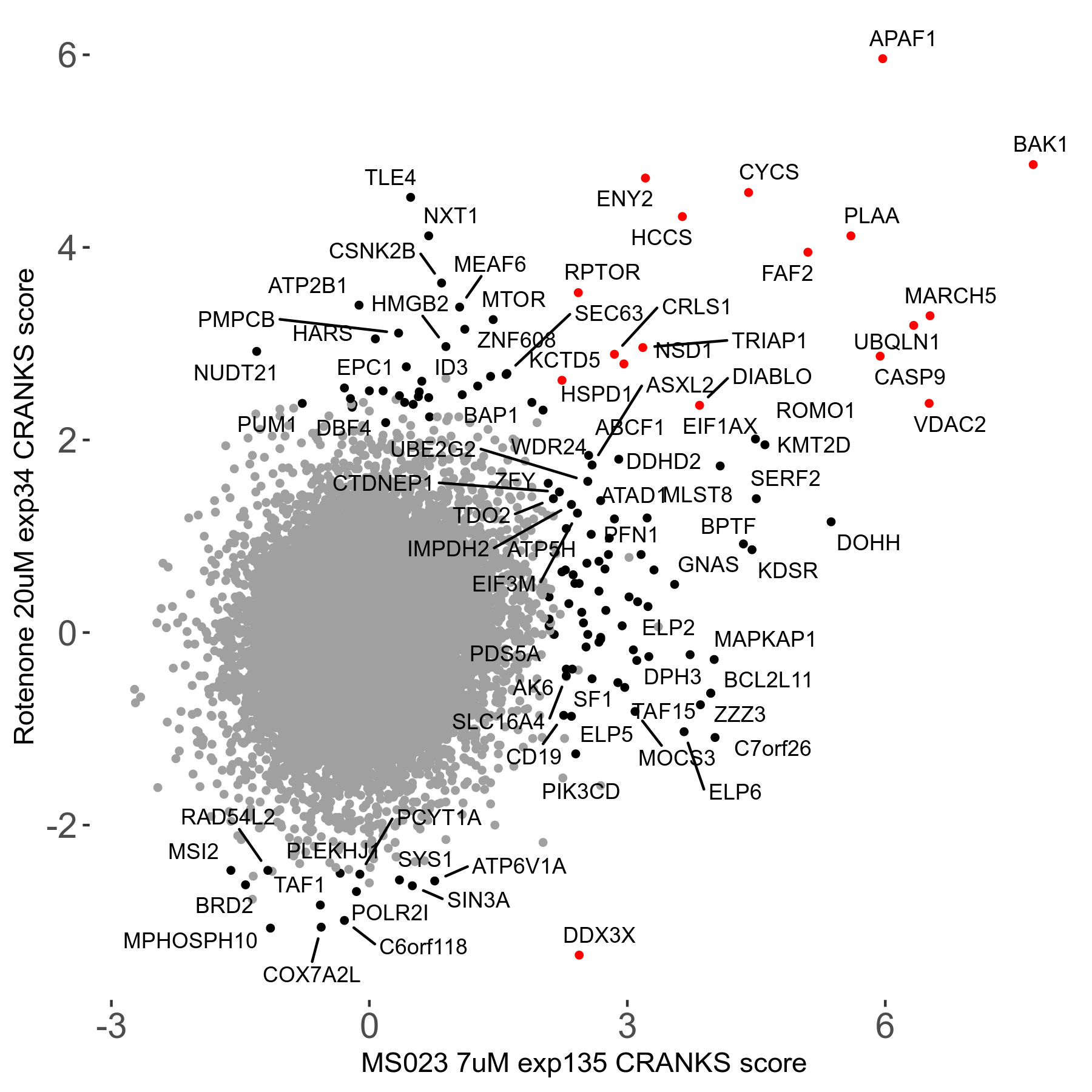
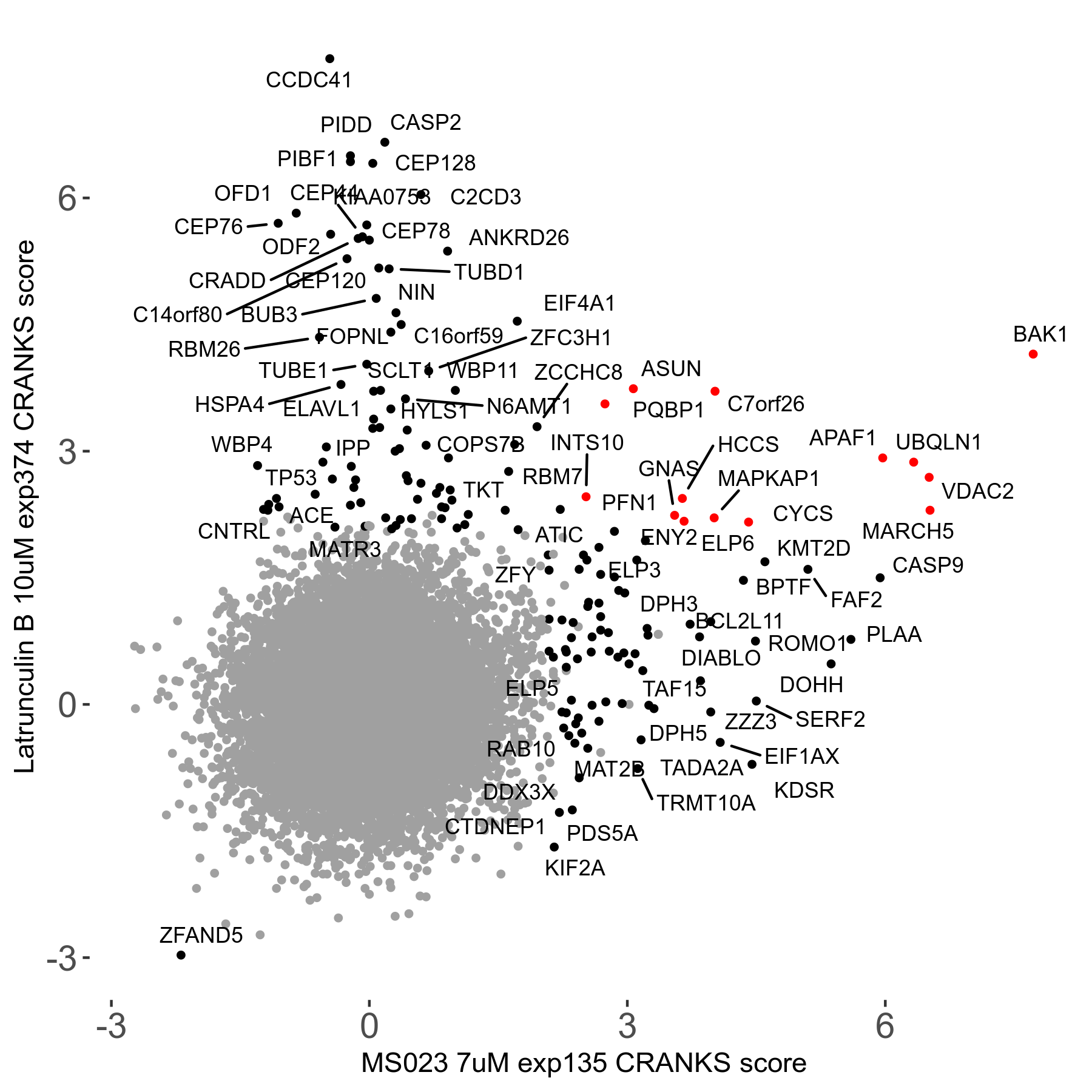
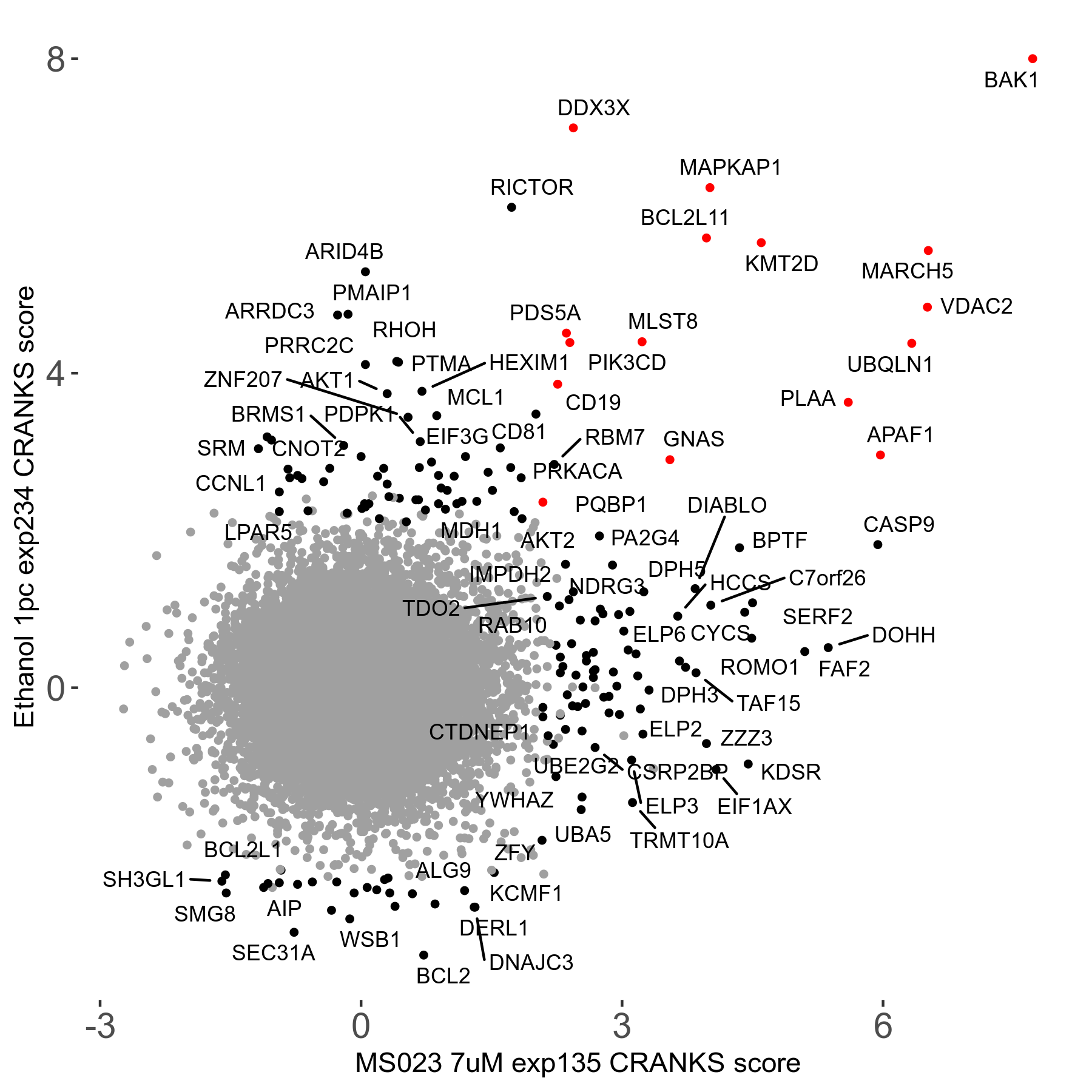
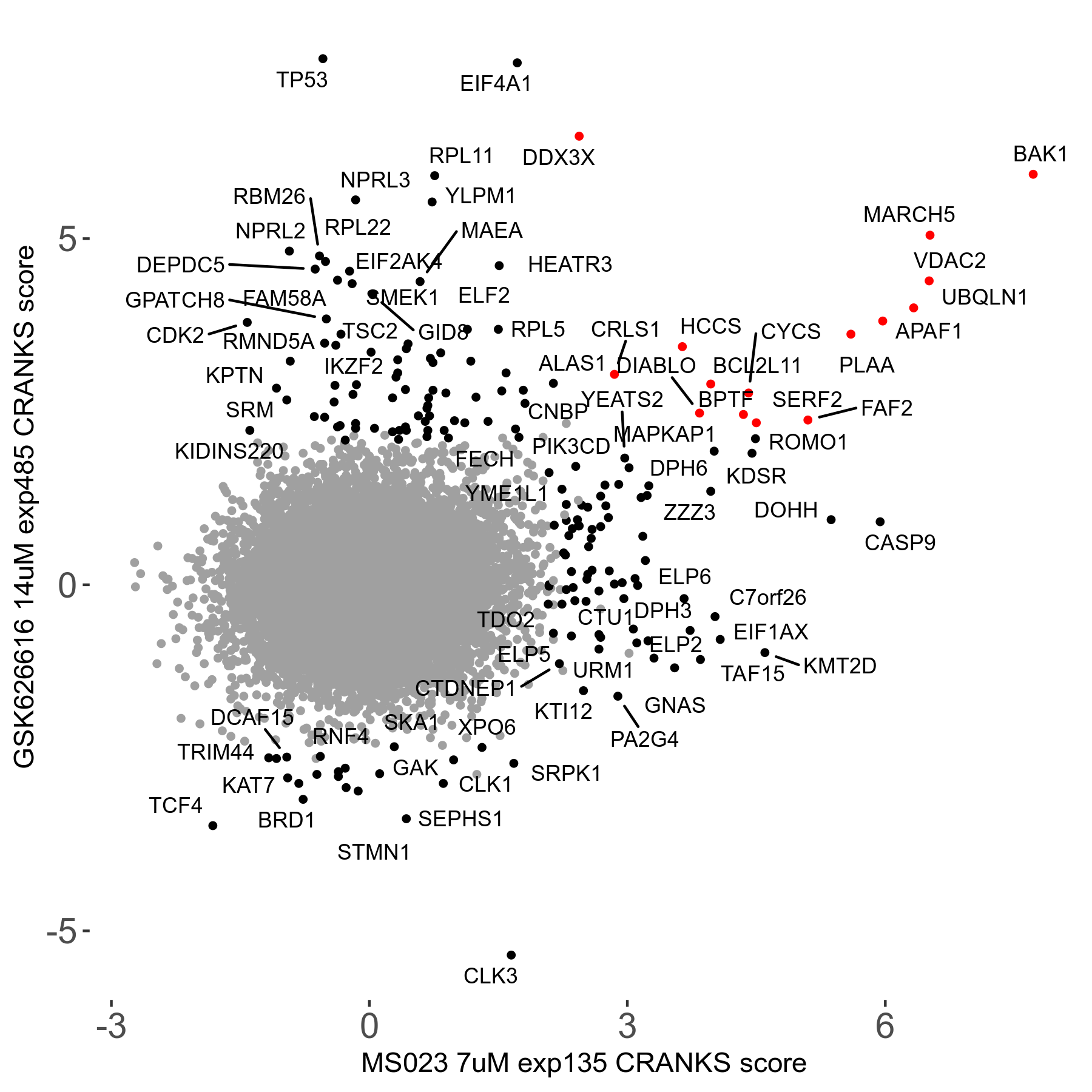
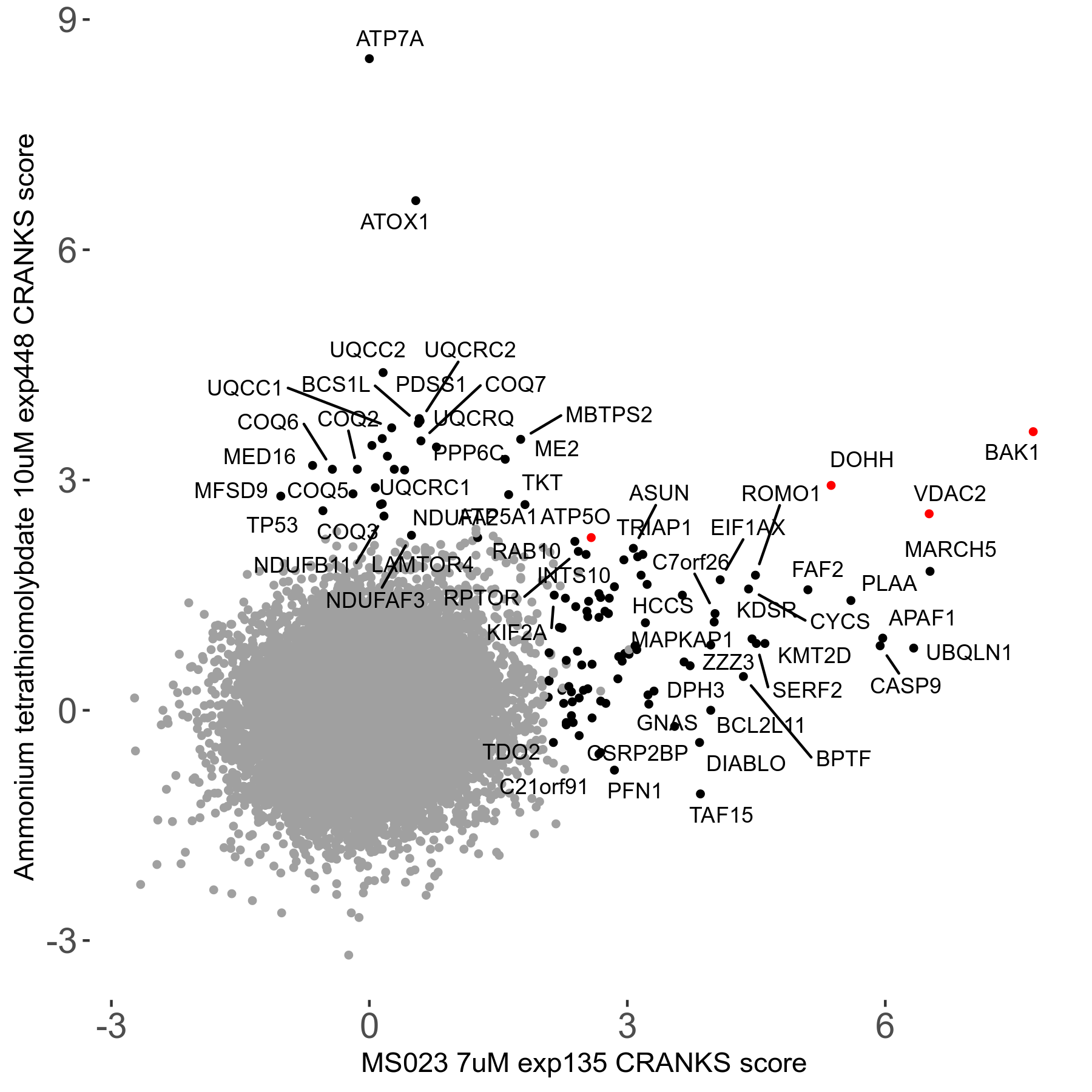
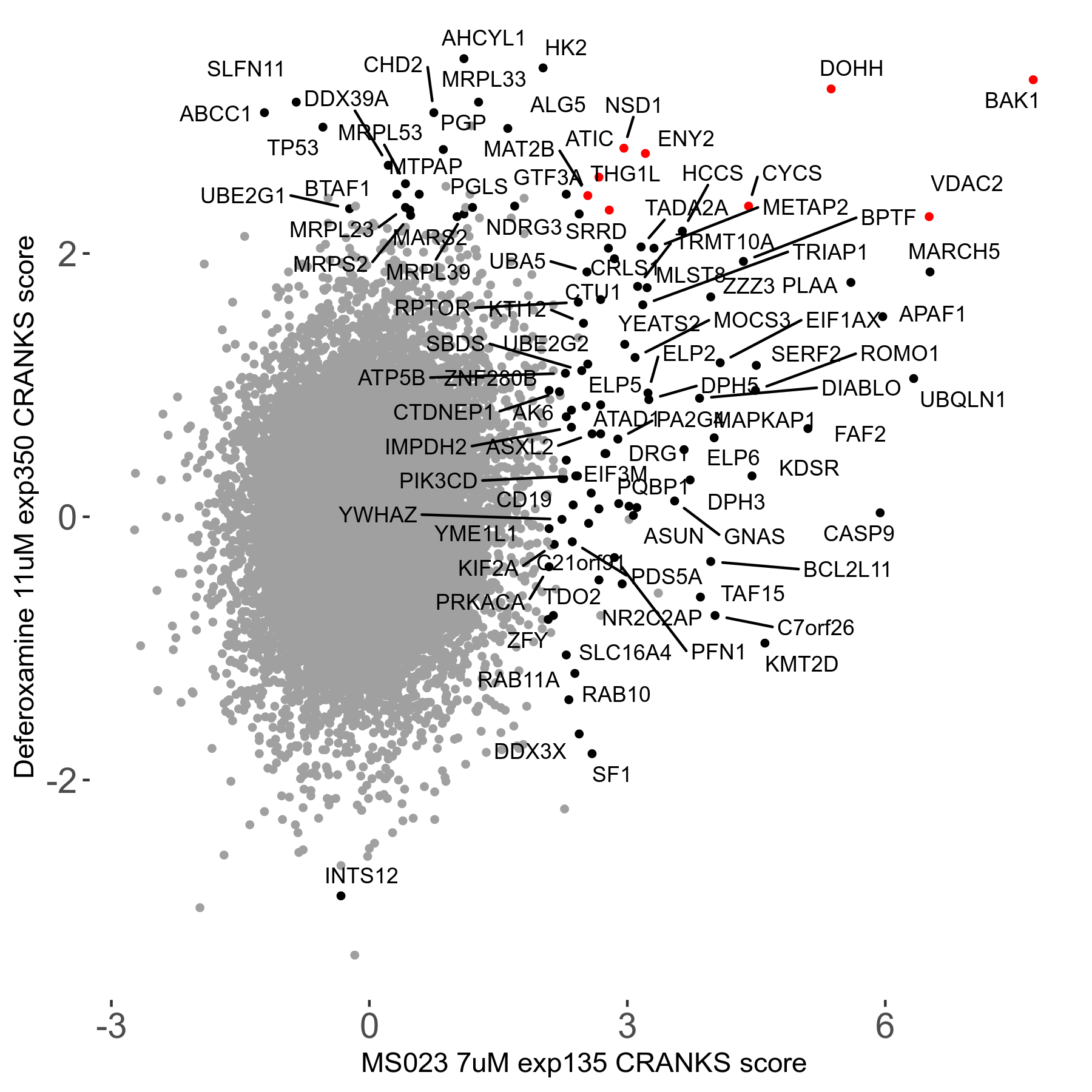
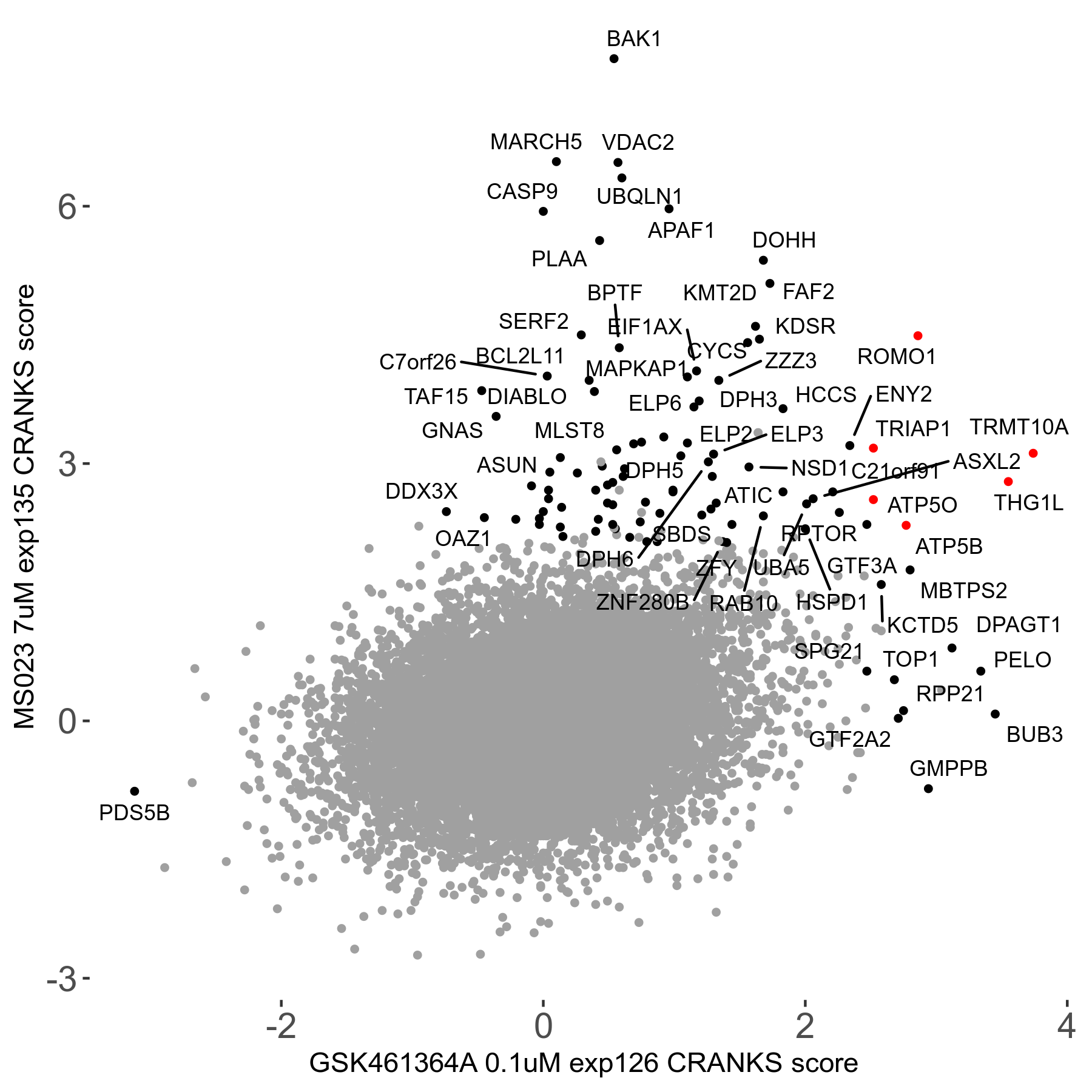
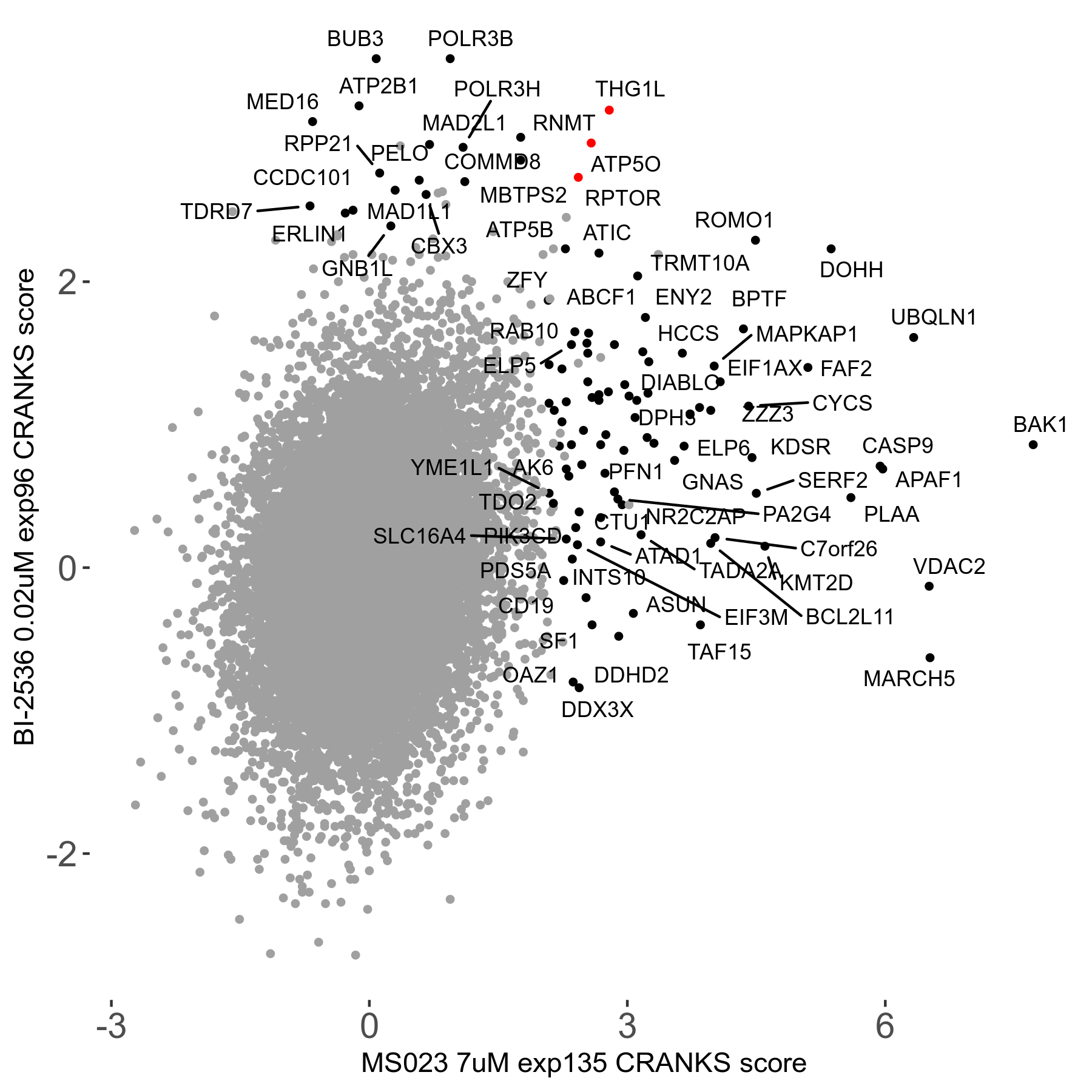
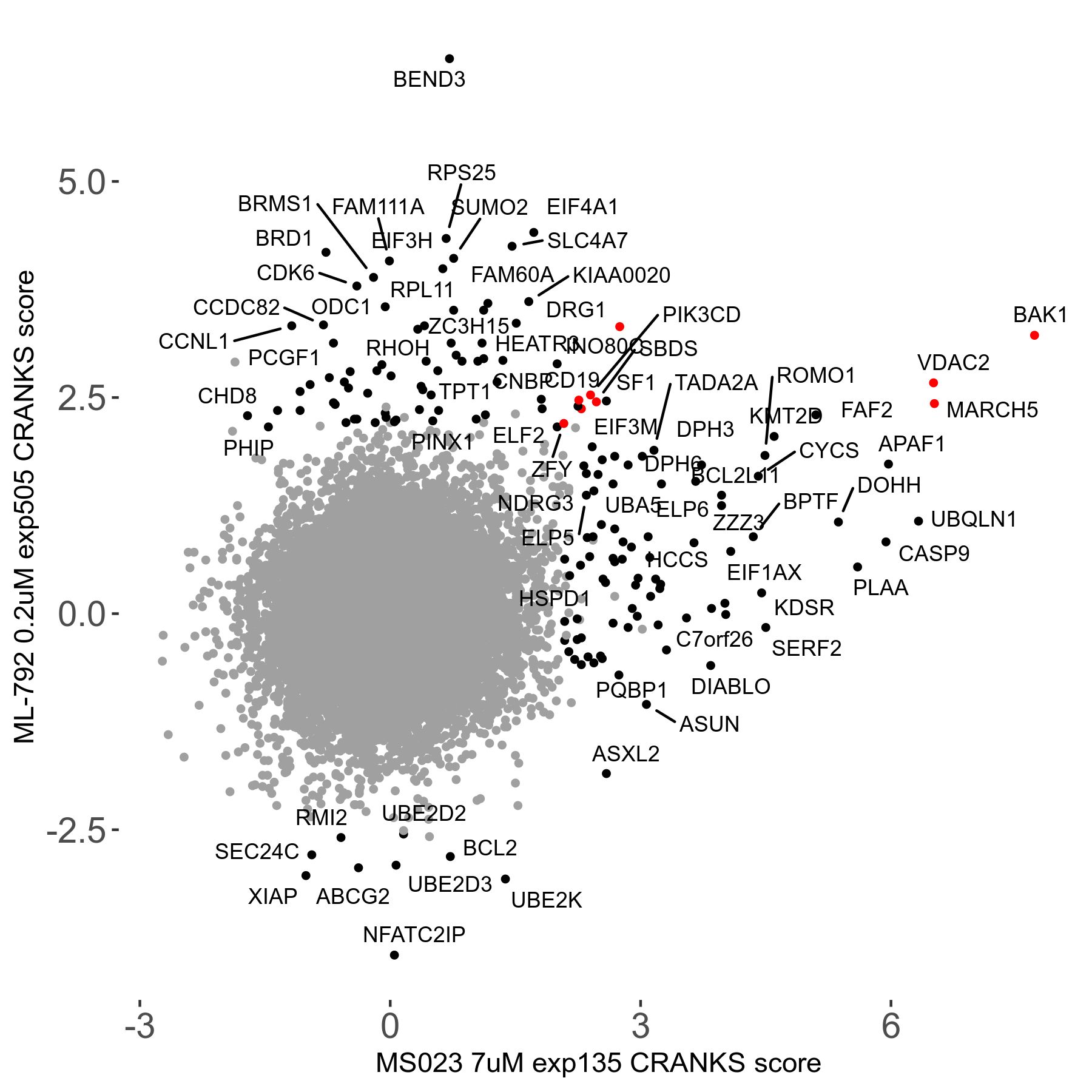
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 0/90 | Scores |