Table of Contents
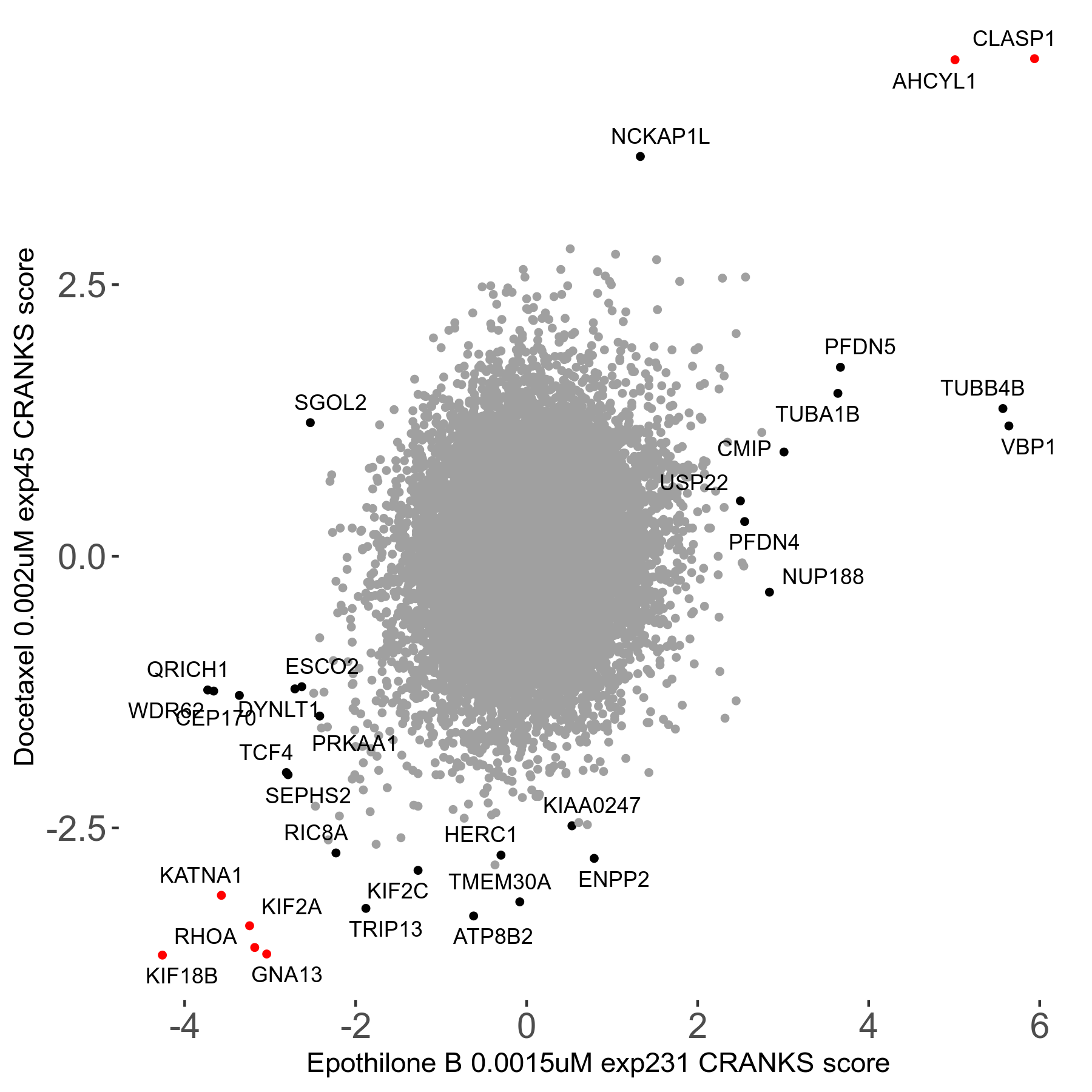
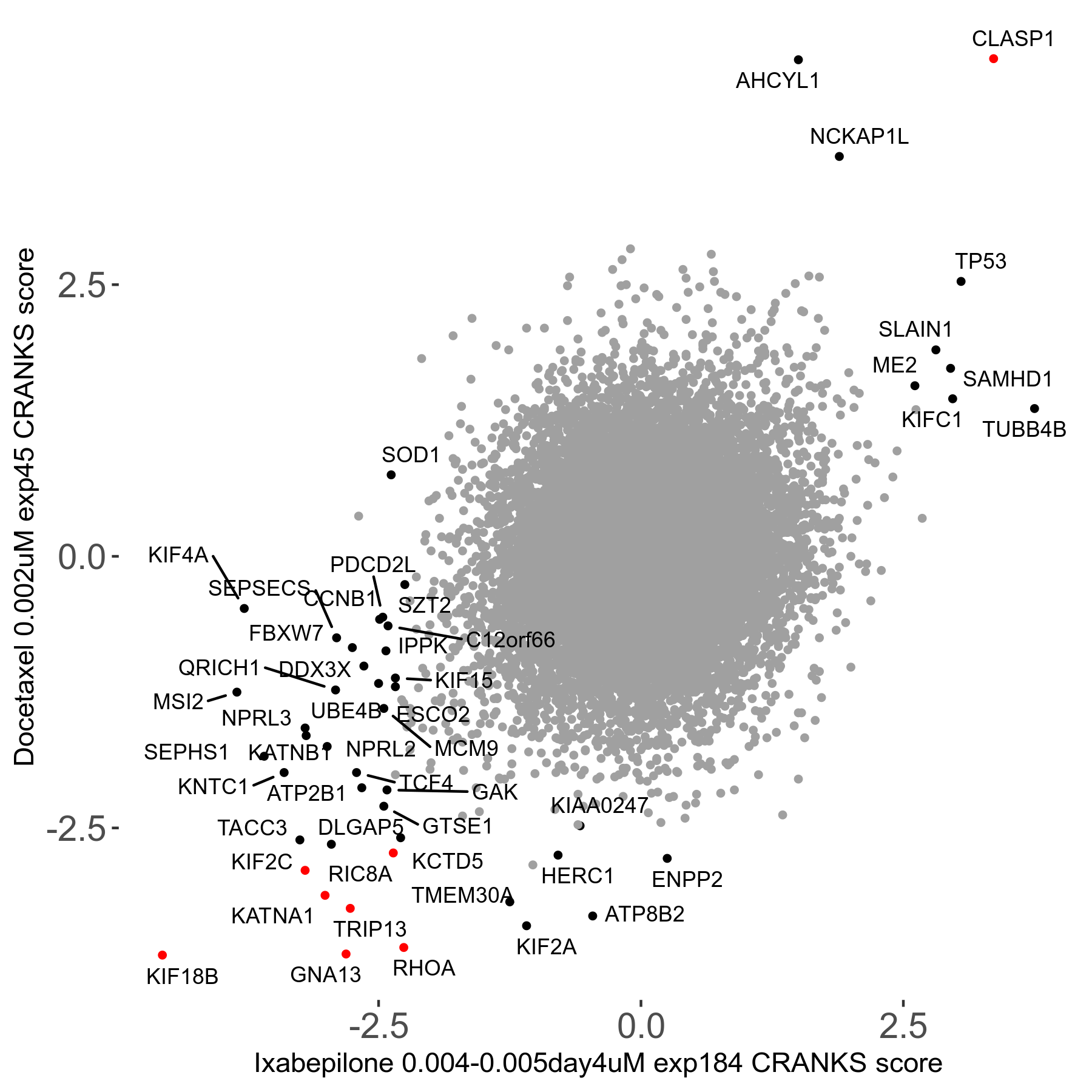
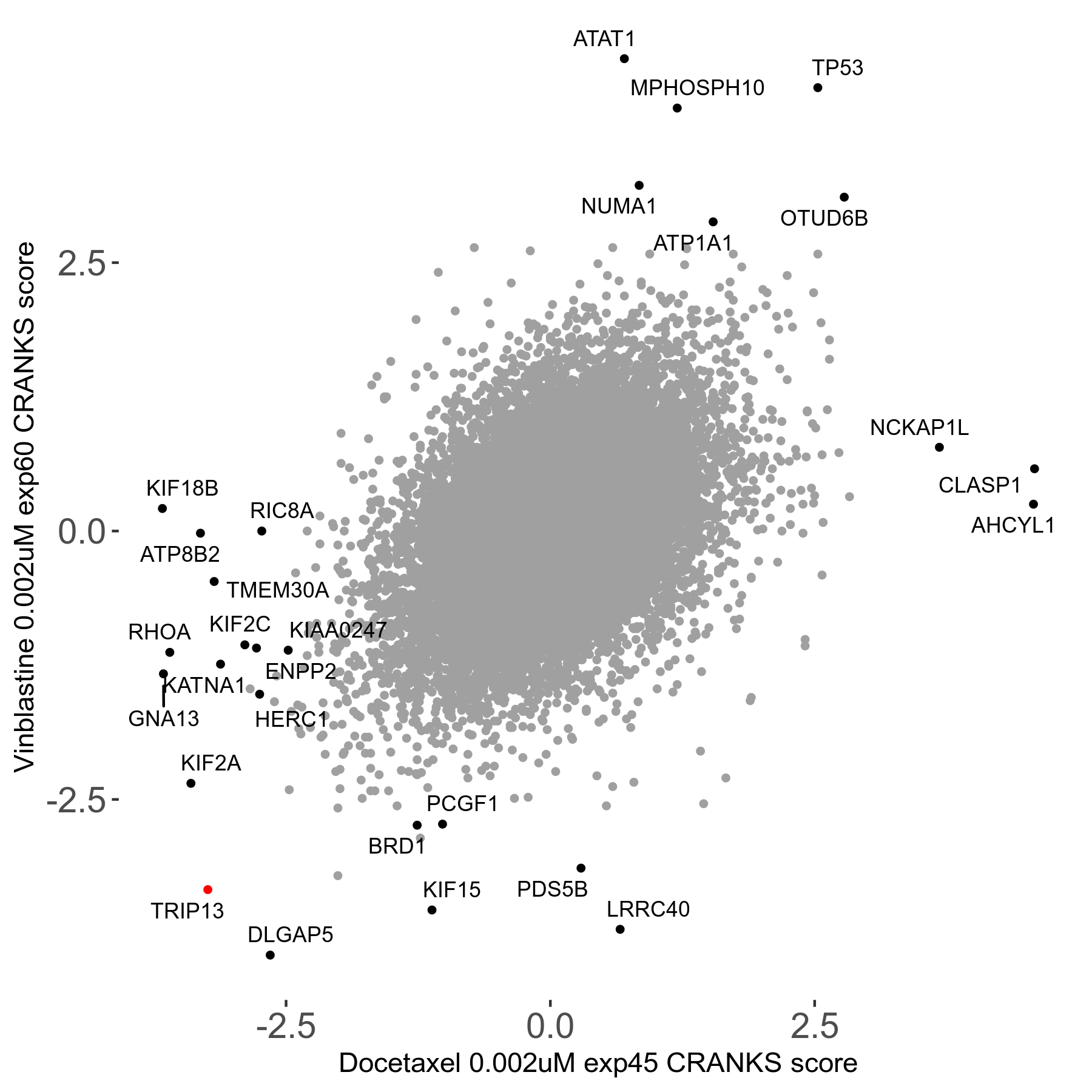
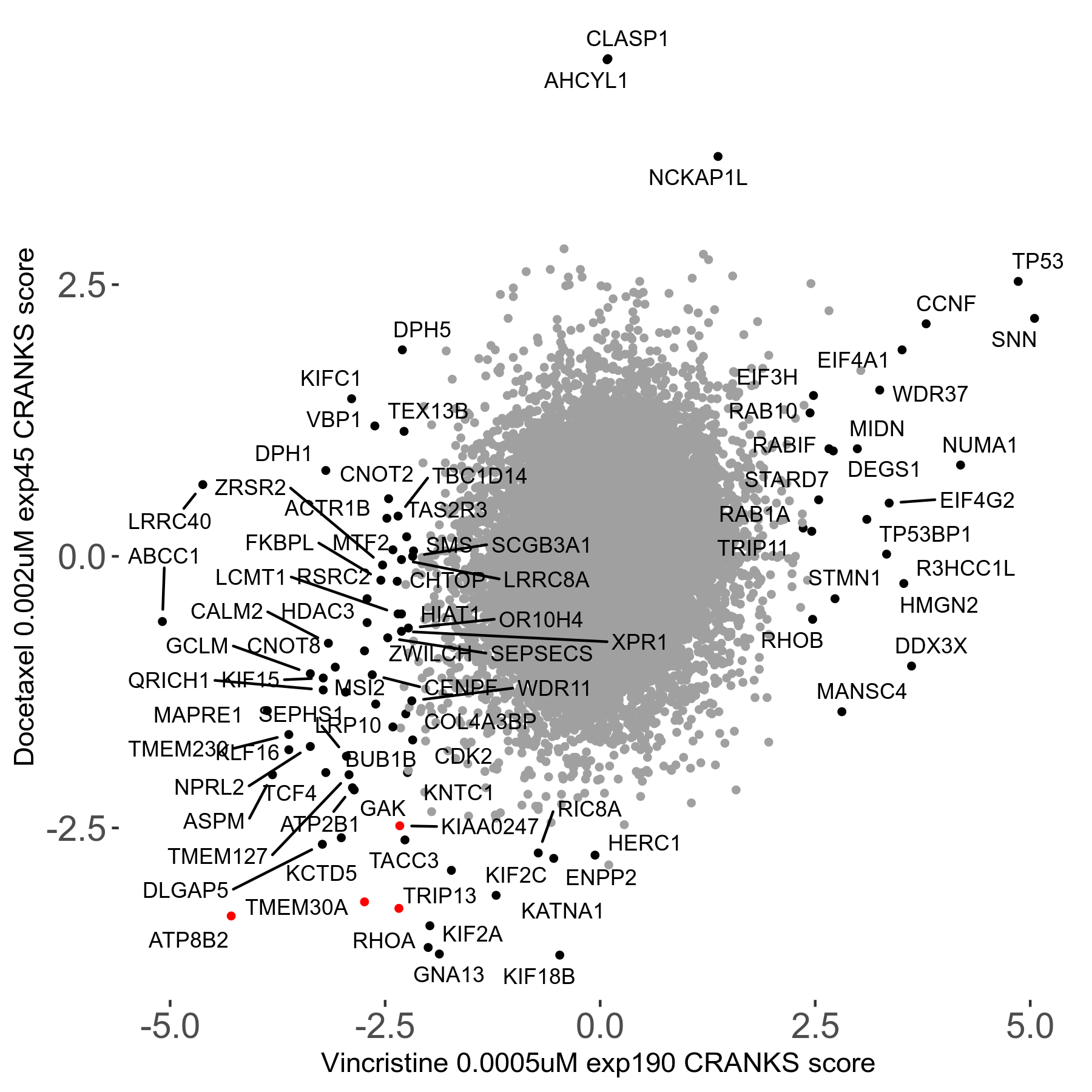
Docetaxel 0.002μM R01 exp45
Mechanism of Action
Inihibts microtubule depolymerization, semi-synthetic analog of the paclitaxel
- Class / Subclass 1: Cell Cycle / Microtubule Poison
- Class / Subclass 2: Organelle Function / Cytoskeletal Inhibitor
Technical Notes
Protein References
- PubChem Name: Docetaxel
- Synonyms: RP-56976
- CAS #: 114977-28-5
- PubChem CID: 148124
- IUPAC: [(1S,2S,3R,4S,7R,9S,10S,12R,15S)-4-acetyloxy-1,9,12-trihydroxy-15-[(2R,3S)-2-hydroxy-3-[(2-methylpropan-2-yl)oxycarbonylamino]-3-phenylpropanoyl]oxy-10,14,17,17-tetramethyl-11-oxo-6-oxatetracyclo[11.3.1.03,10.04,7]heptadec-13-en-2-yl] benzoate
- INCHI Name: InChI=1S/C43H53NO14/c1-22-26(55-37(51)32(48)30(24-15-11-9-12-16-24)44-38(52)58-39(3,4)5)20-43(53)35(56-36(50)25-17-13-10-14-18-25)33-41(8,34(49)31(47)29(22)40(43,6)7)27(46)19-28-42(33,21-54-28)57-23(2)45/h9-18,26-28,30-33,35,46-48,53H,19-21H2,1-8H3,(H,44,52)/t26-,27-,28+,30-,31+,32+,33-,35-,41+,42-,43+/m0/s1
- INCHI Key: ZDZOTLJHXYCWBA-VCVYQWHSSA-N
- Molecular Weight: 807.9
- Canonical SMILES: CC1=C2C(C(=O)C3(C(CC4C(C3C(C(C2(C)C)(CC1OC(=O)C(C(C5=CC=CC=C5)NC(=O)OC(C)(C)C)O)O)OC(=O)C6=CC=CC=C6)(CO4)OC(=O)C)O)C)O
- Isomeric SMILES: CC1=C2[C@H](C(=O)[C@@]3([C@H](C[C@@H]4[C@]([C@H]3[C@@H]([C@@](C2(C)C)(C[C@@H]1OC(=O)[C@@H]([C@H](C5=CC=CC=C5)NC(=O)OC(C)(C)C)O)O)OC(=O)C6=CC=CC=C6)(CO4)OC(=O)C)O)C)O
- Molecular Formula: C43H53NO14
Protein Supplier
- Supplier Name: Abcam
- Catalog #: AB141248
- Lot #: N/A
Protein Characterization
- HRMS (ESI-TOF) m/z: (M+Na)+ Calcd for C43H53NO14 830.33583; found 830.33216
Dose Response Curve
- Platform ID: DOCETAXEL
- Min: 6.3537; Max: 75.1022

| IC | Concentration (µM) |
|---|---|
| IC10 | N/A |
| IC20 | N/A |
| IC30 | 0.0002 |
| IC40 | 0.0006 |
| IC50 | 0.0020 |
| IC60 | N/A |
| IC70 | N/A |
| IC80 | N/A |
| IC90 | N/A |
Screen Summary
- Round: 01
- Dose: 2nM
- Days of incubation: 8
- Doublings: 5.2
- Numbers of reads: 15167862
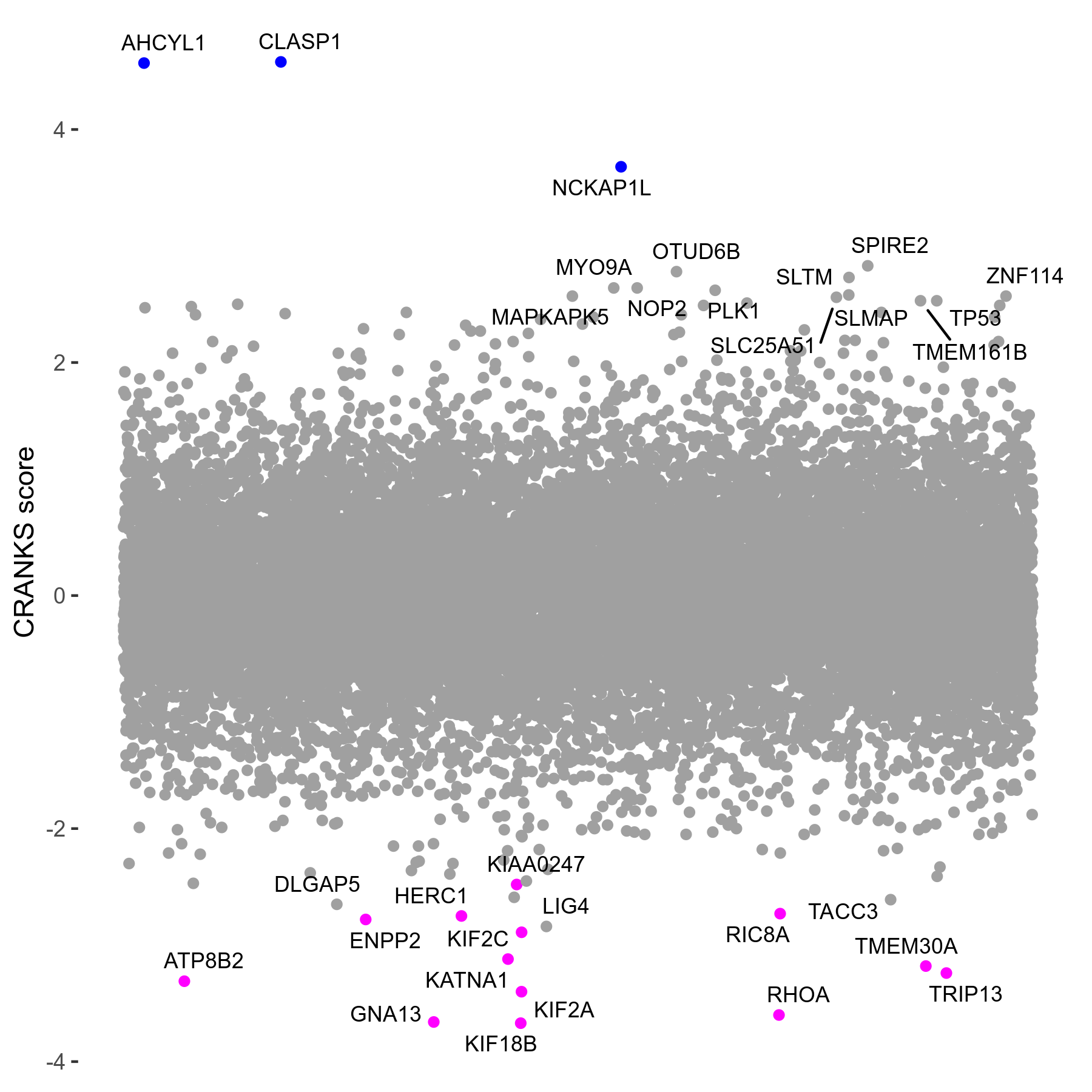
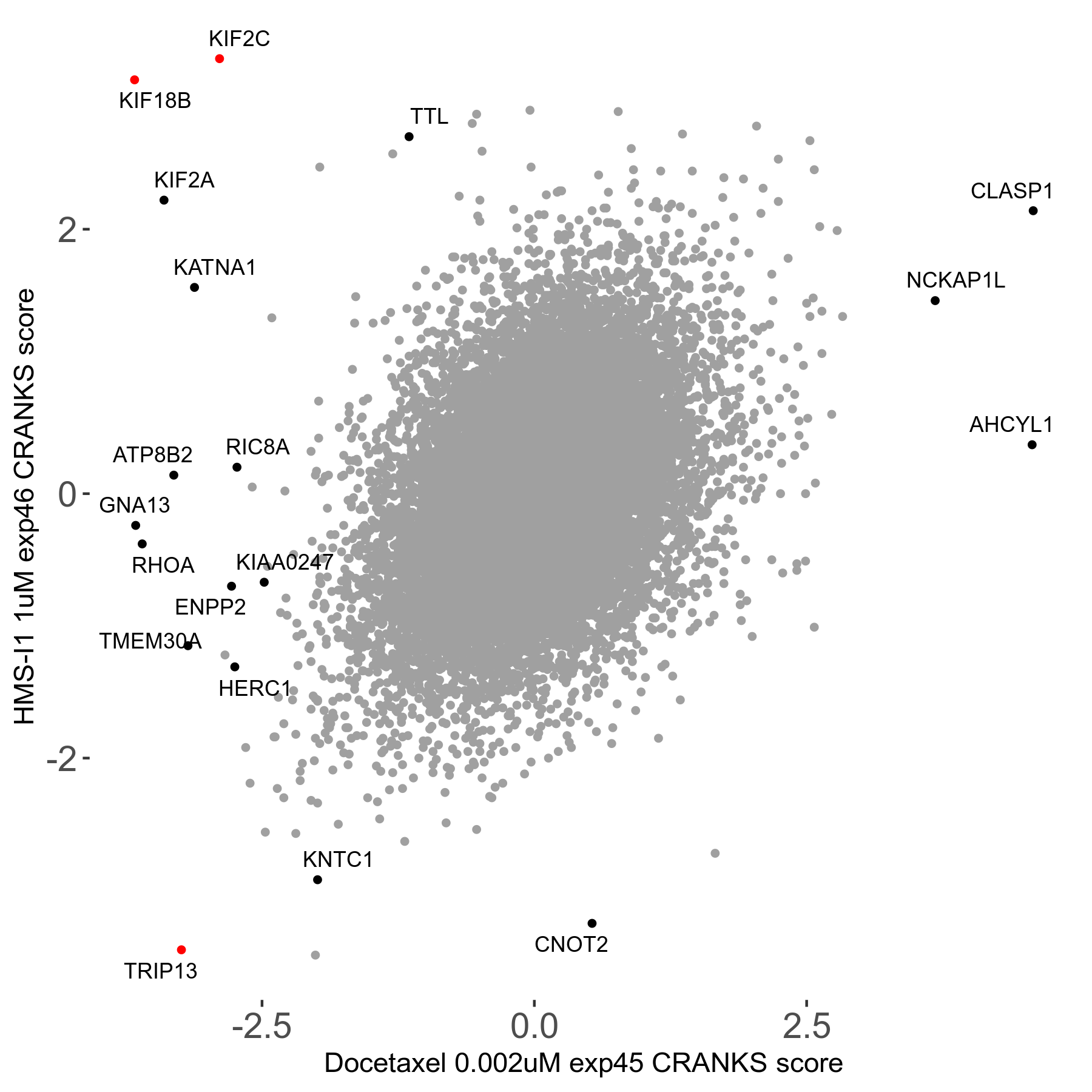
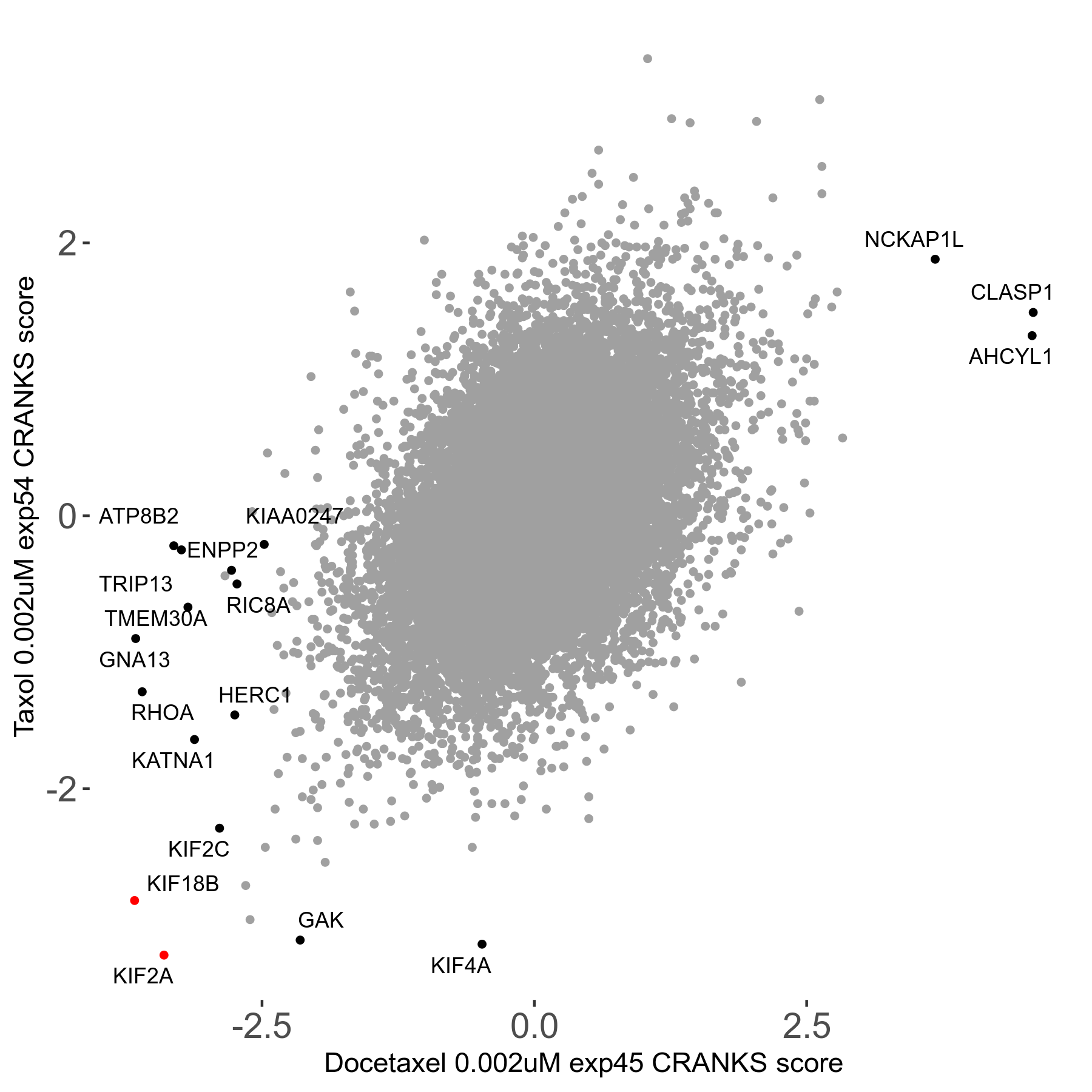
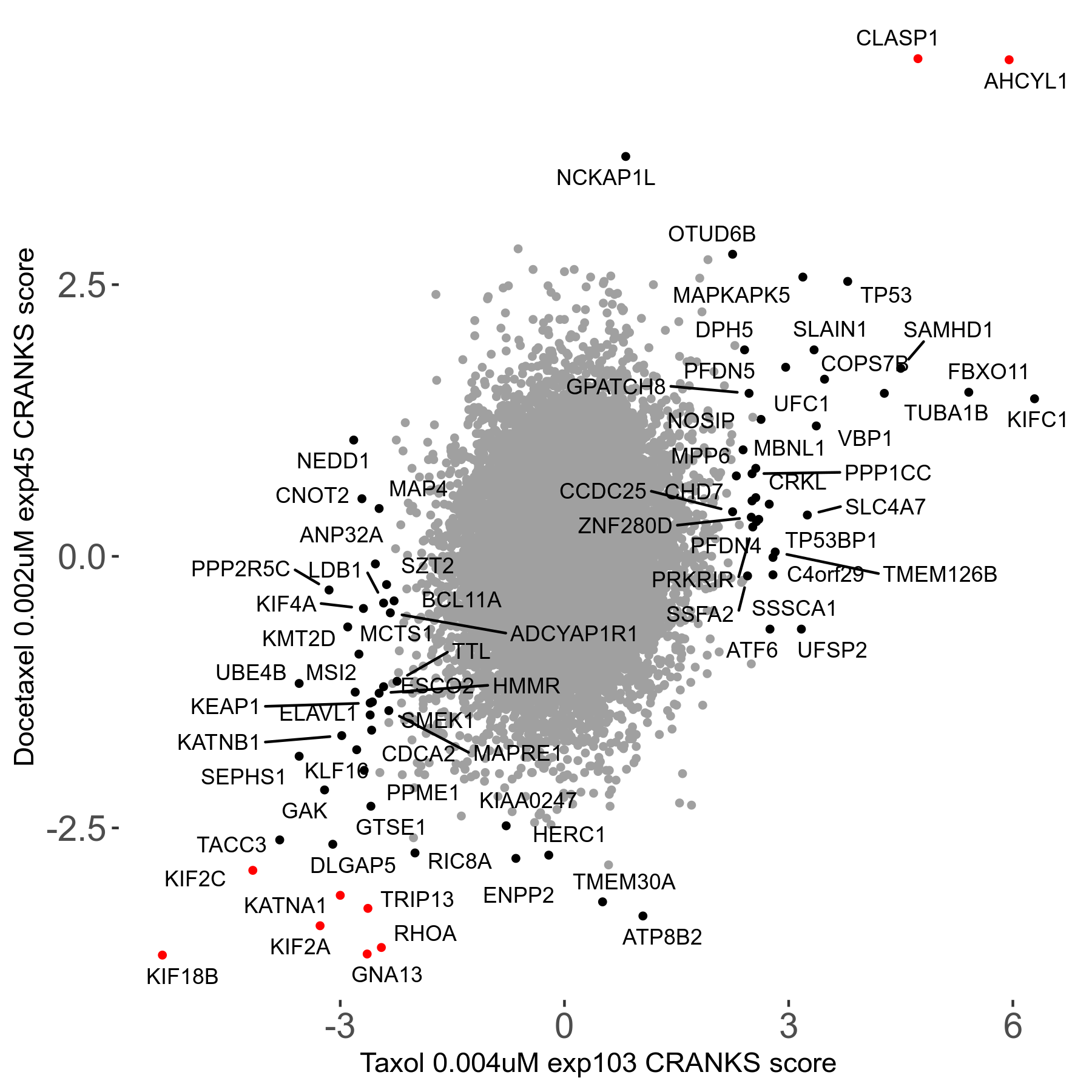
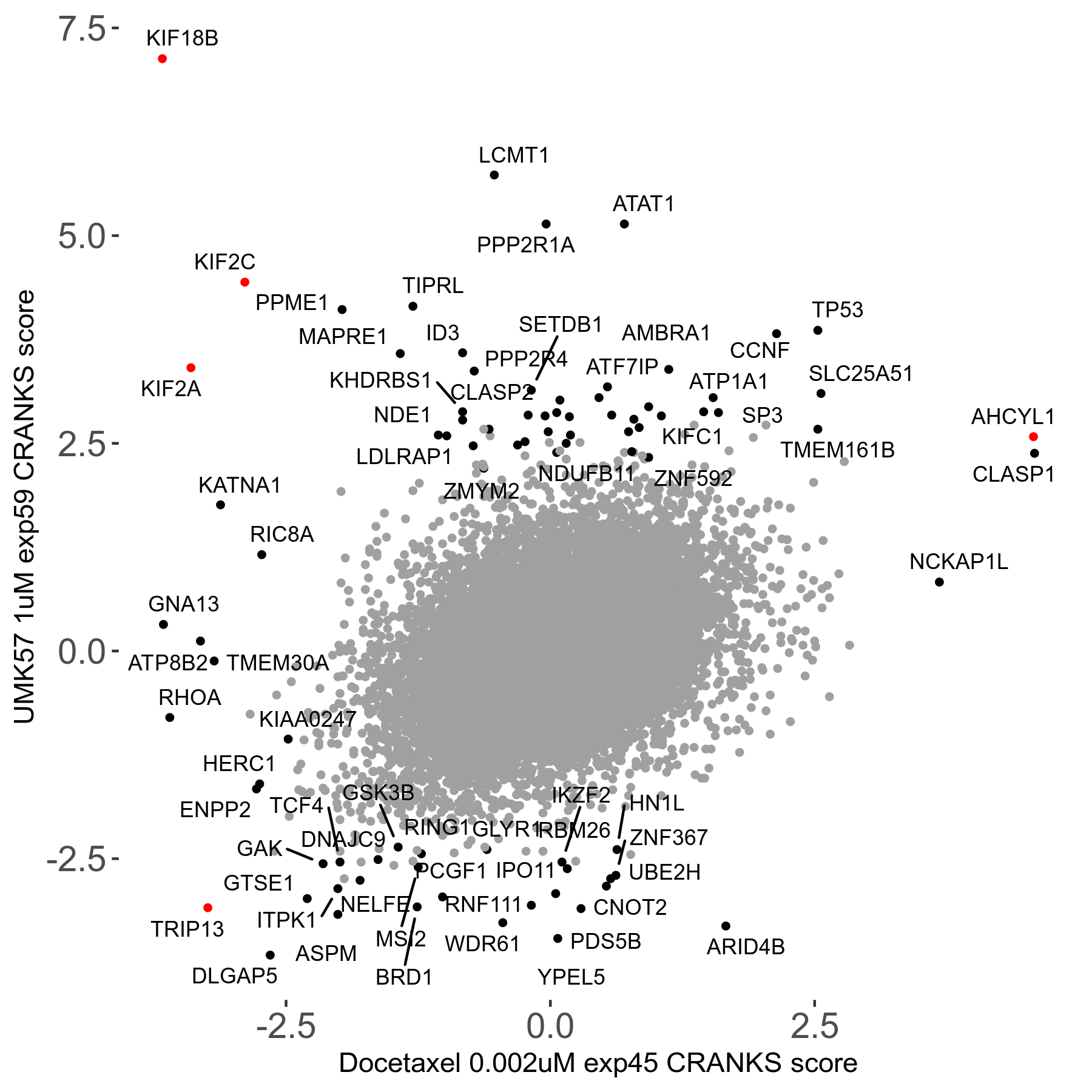
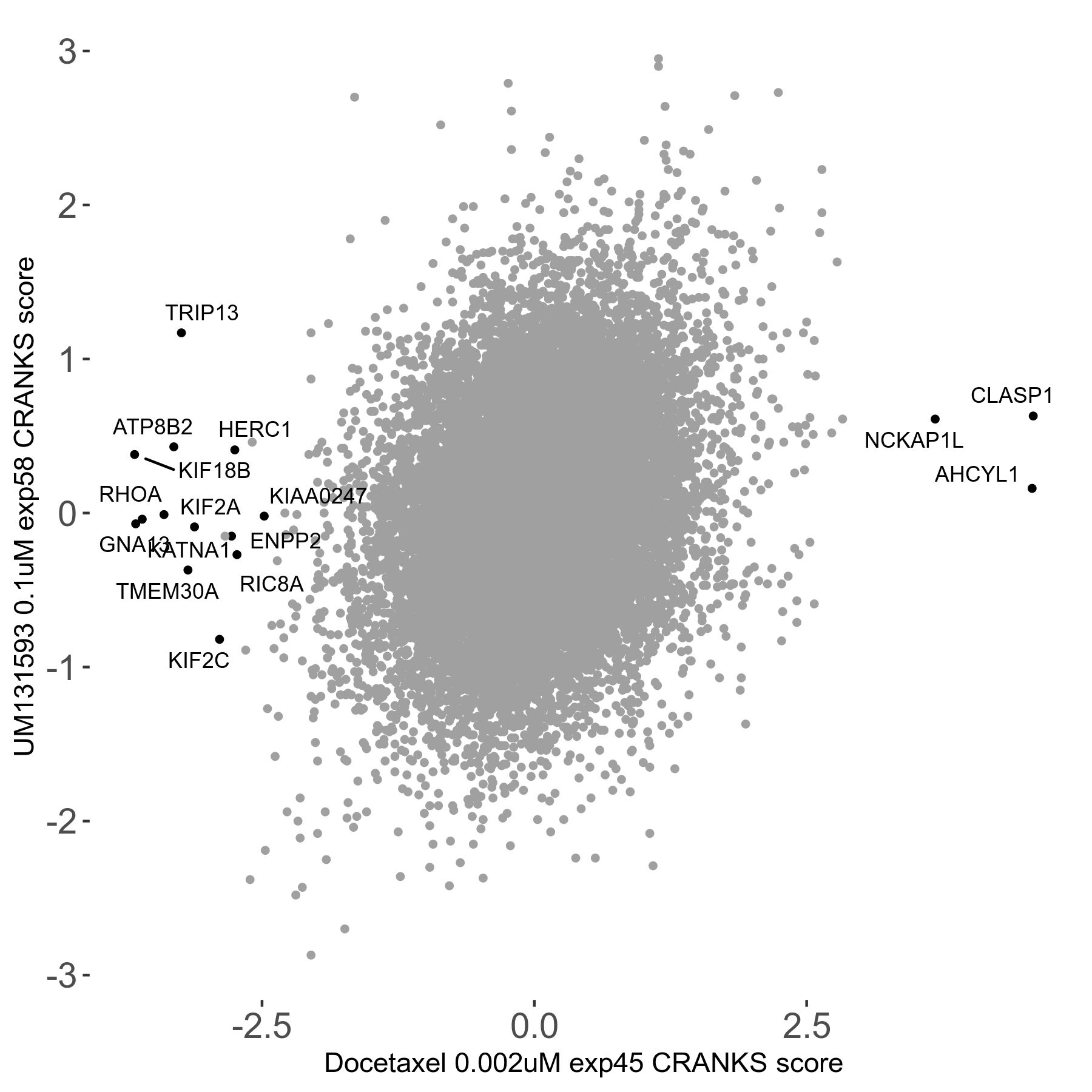
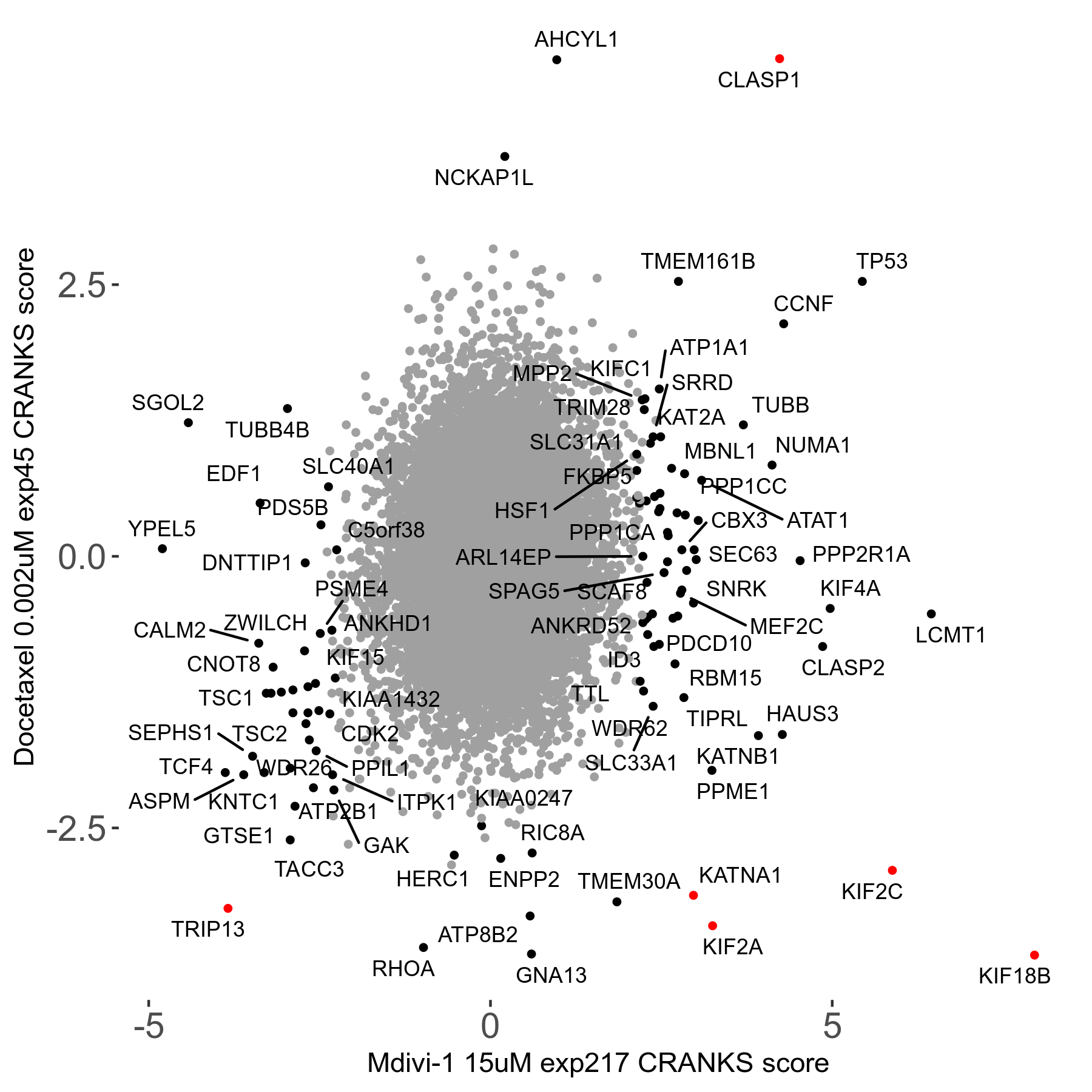
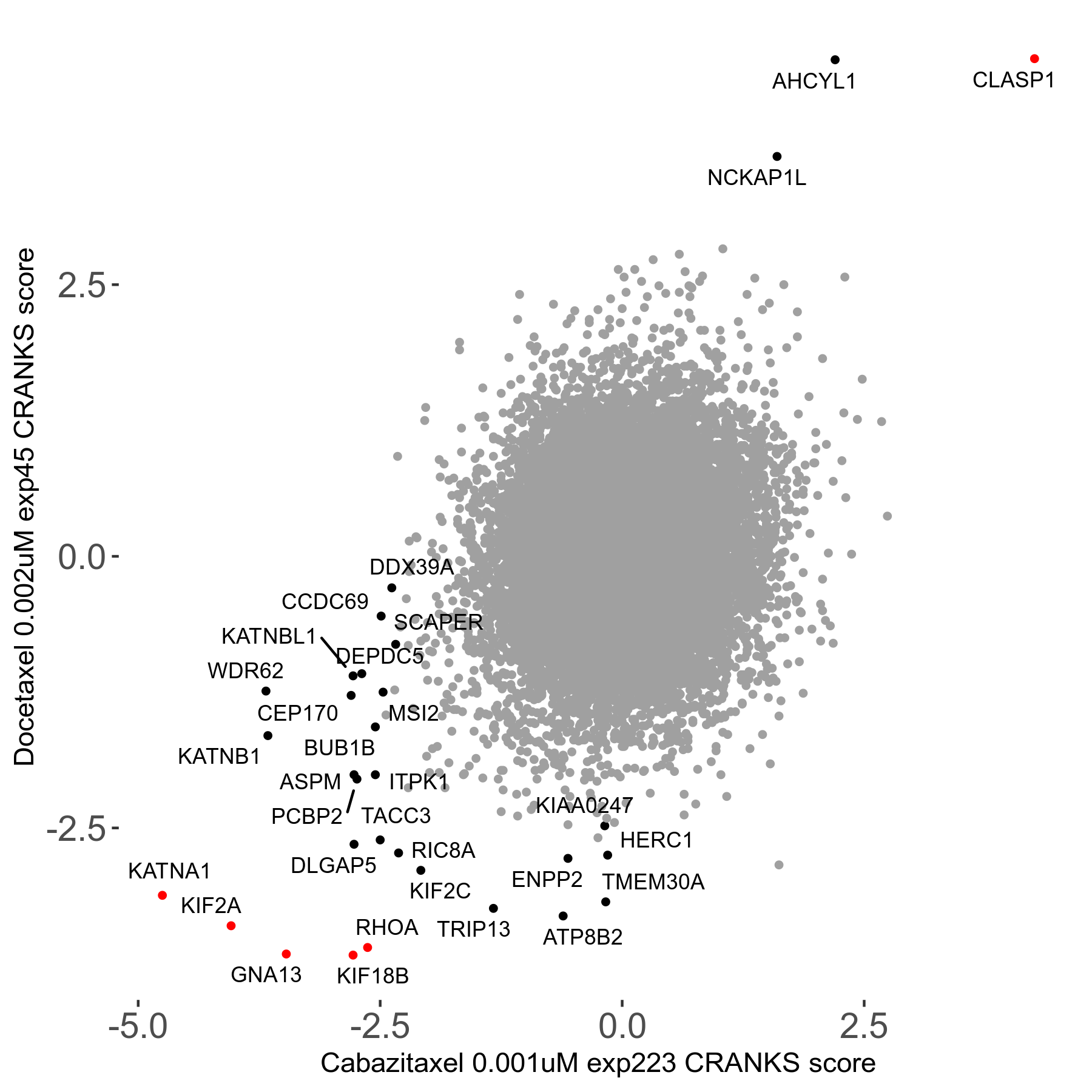
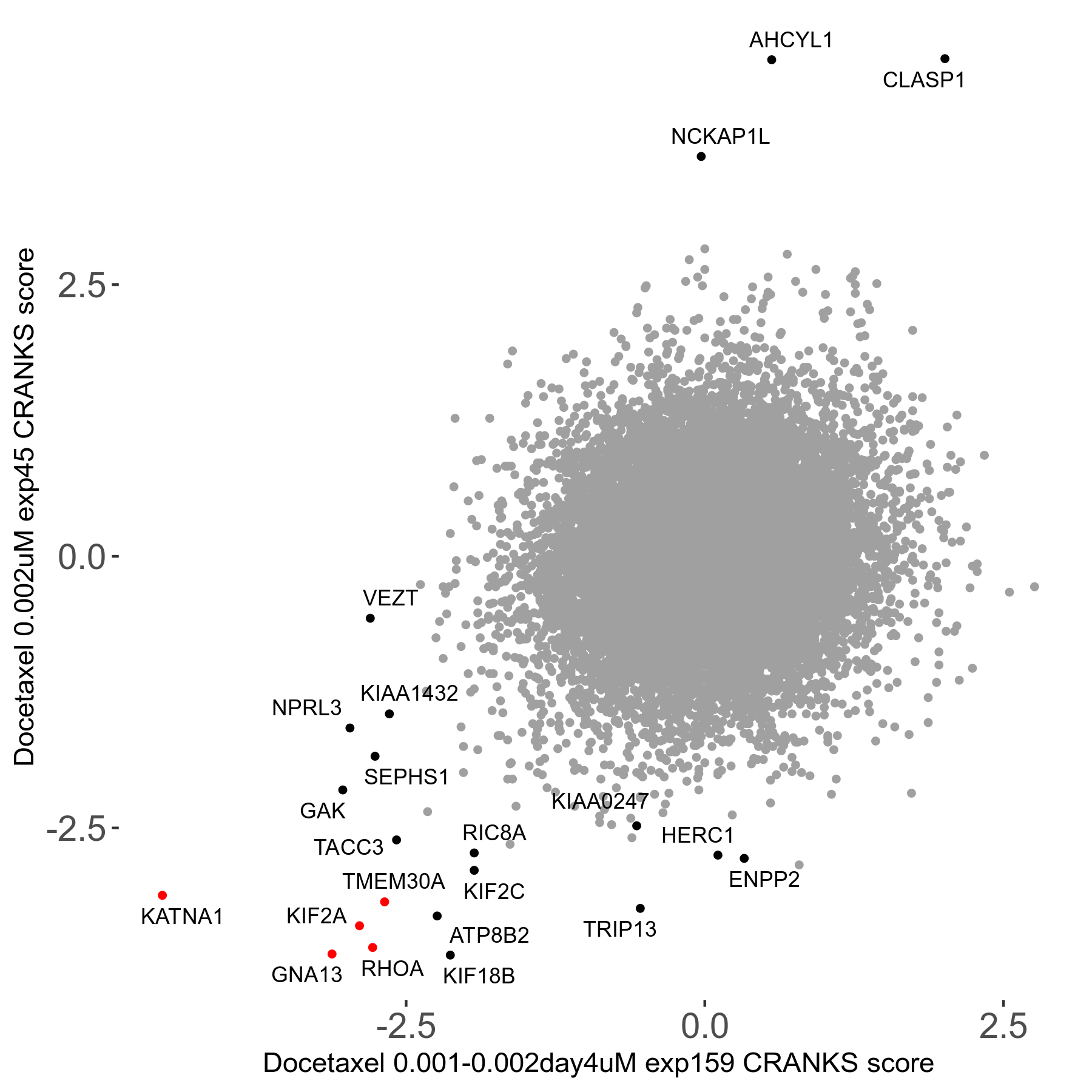
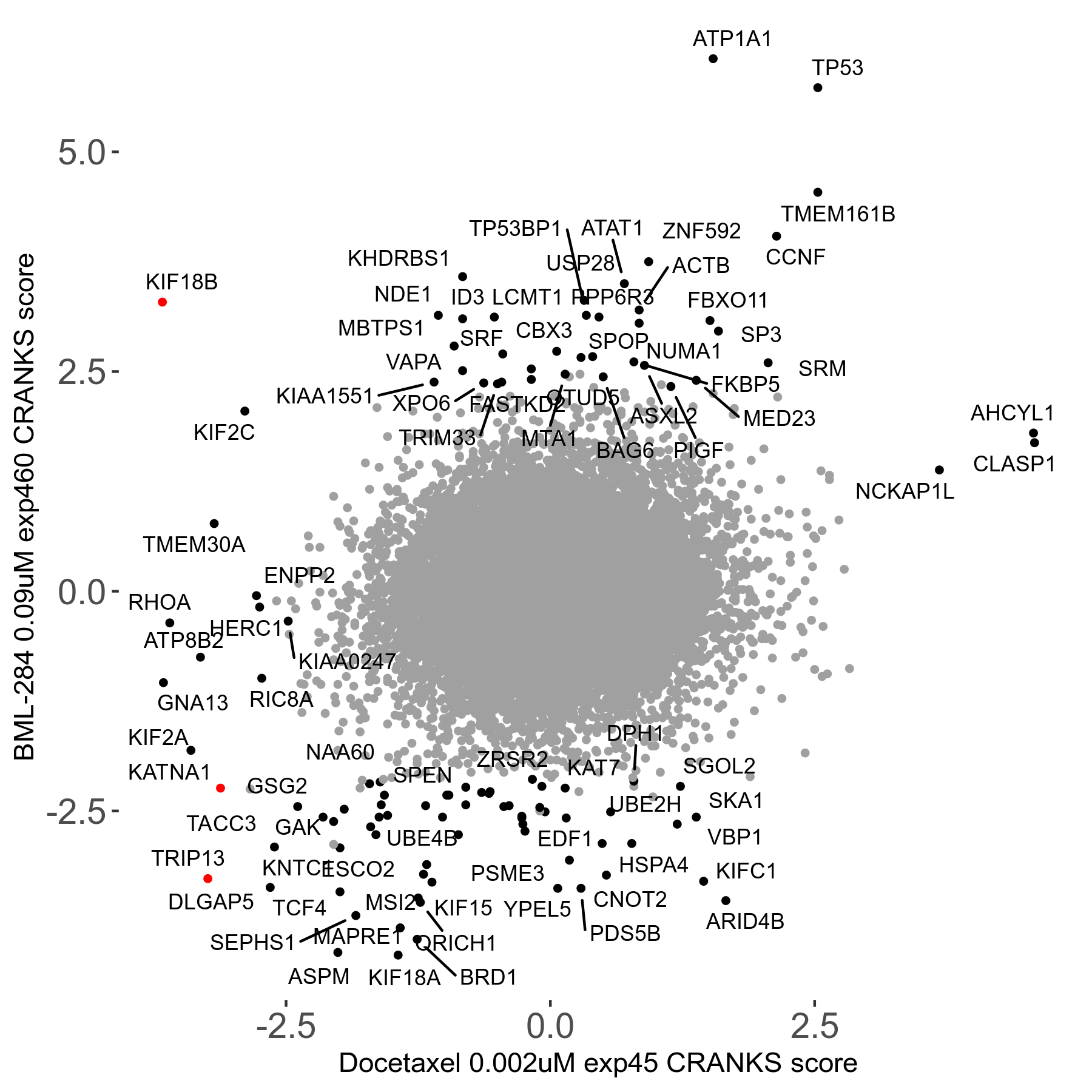
Screen Results
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 13/3 | Scores |