42°C R08 exp539
Mechanism of Action
Elevated growth temperature
- Class / Subclass 1: Environmental Stresses / Stress Condition
Screen Summary
- Round: 08
- Dose: N/A
- Days of incubation: 8
- Doublings: -0.5
- Numbers of reads: 18691098
Screen Results
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 1/26 | Scores | View | View | View | View | View |
42°C R08 exp539
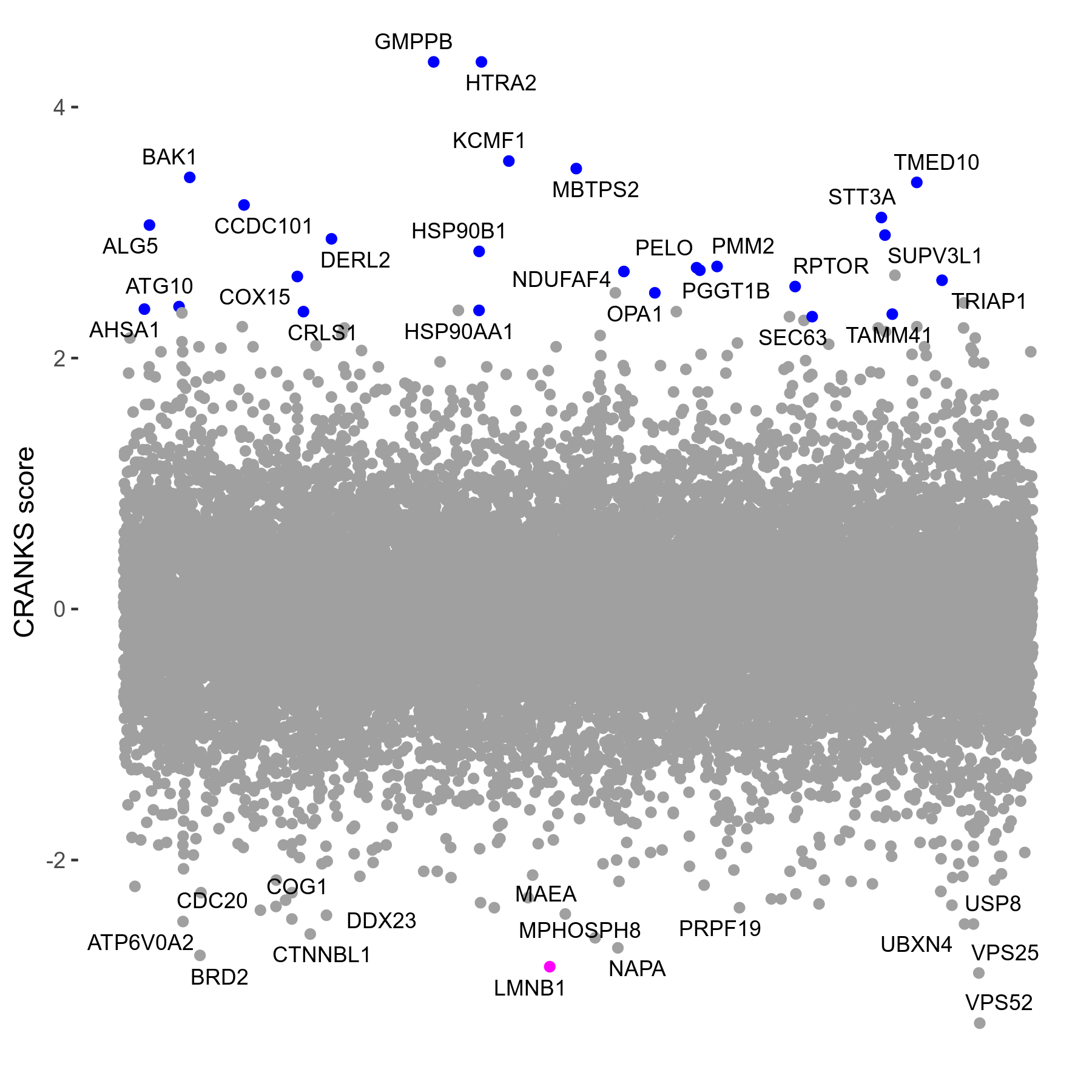
Top 30 S/R Genes
| Gene | CRANKS Score | FDR |
|---|---|---|
| VPS52 | -3.30 | 0.07 |
| VPS25 | -2.90 | 0.19 |
| LMNB1 | -2.85 | 0.05 |
| BRD2 | -2.76 | 0.06 |
| NAPA | -2.70 | 0.17 |
| MPHOSPH8 | -2.62 | 0.06 |
| CTNNBL1 | -2.59 | 0.06 |
| USP8 | -2.51 | 0.12 |
| UBXN4 | -2.51 | 0.07 |
| ATP6V0A2 | -2.49 | 0.10 |
| COG1 | -2.47 | 0.07 |
| DDX23 | -2.44 | 0.36 |
| MAEA | -2.43 | 0.07 |
| CDC20 | -2.40 | 0.19 |
| PRPF19 | -2.38 | 0.20 |
| ILF3 | -2.38 | 0.27 |
| CHMP2A | -2.37 | 0.20 |
| TSSC1 | -2.36 | 0.10 |
| SF3A1 | -2.35 | 0.20 |
| HTATSF1 | -2.34 | 0.17 |
| CLN3 | -2.32 | 0.12 |
| RCN1 | -2.31 | 0.11 |
| RINT1 | -2.31 | 0.17 |
| KMT2E | -2.30 | 0.11 |
| RRM2 | -2.27 | 0.52 |
| COG3 | -2.26 | 0.11 |
| BROX | -2.26 | 0.11 |
| TRAPPC13 | -2.25 | 0.11 |
| TRAPPC2L | -2.25 | 0.11 |
| ACYP2 | -2.21 | 0.16 |
| ATP5A1 | 2.36 | 0.09 |
| CRLS1 | 2.37 | 0.04 |
| OTUD5 | 2.37 | 0.09 |
| HSP90AA1 | 2.38 | 0.04 |
| HCCS | 2.38 | 0.09 |
| AHSA1 | 2.39 | 0.04 |
| ATG10 | 2.41 | 0.04 |
| UBR4 | 2.44 | 0.08 |
| N6AMT1 | 2.52 | 0.09 |
| OPA1 | 2.52 | 0.04 |
| RPTOR | 2.57 | 0.04 |
| TRIAP1 | 2.62 | 0.04 |
| COX15 | 2.65 | 0.01 |
| TAZ | 2.66 | 0.12 |
| NDUFAF4 | 2.69 | <0.01 |
| PGGT1B | 2.70 | <0.01 |
| PELO | 2.72 | 0.04 |
| PMM2 | 2.73 | <0.01 |
| HSP90B1 | 2.85 | <0.01 |
| DERL2 | 2.95 | <0.01 |
| SUPV3L1 | 2.98 | 0.01 |
| ALG5 | 3.06 | <0.01 |
| STT3A | 3.12 | 0.02 |
| CCDC101 | 3.22 | <0.01 |
| TMED10 | 3.40 | <0.01 |
| BAK1 | 3.44 | <0.01 |
| MBTPS2 | 3.51 | <0.01 |
| KCMF1 | 3.57 | <0.01 |
| HTRA2 | 4.36 | <0.01 |
| GMPPB | 4.36 | <0.01 |
Correlated Screens
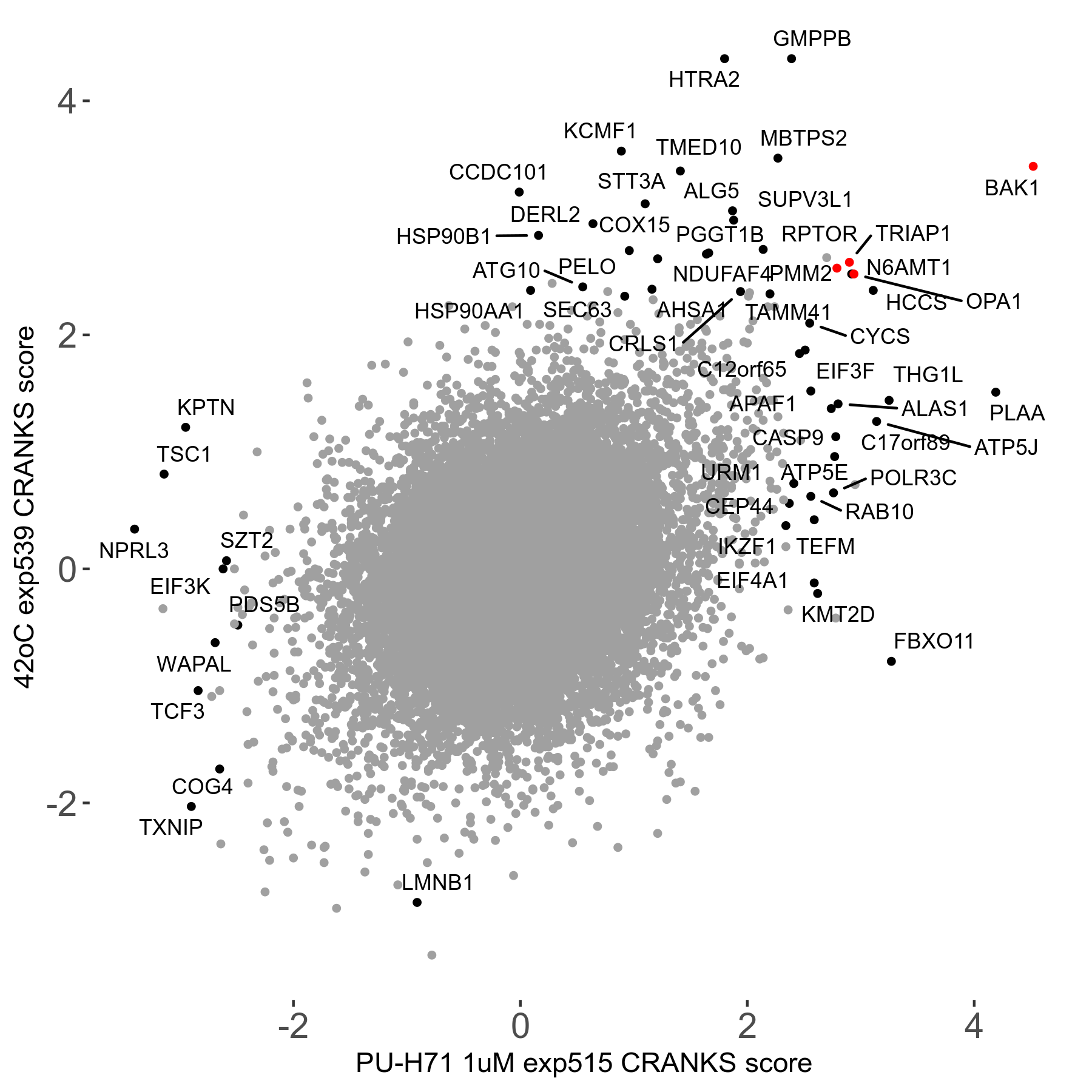
42°C R08 exp539 vs PU-H71 1μM R08 exp515
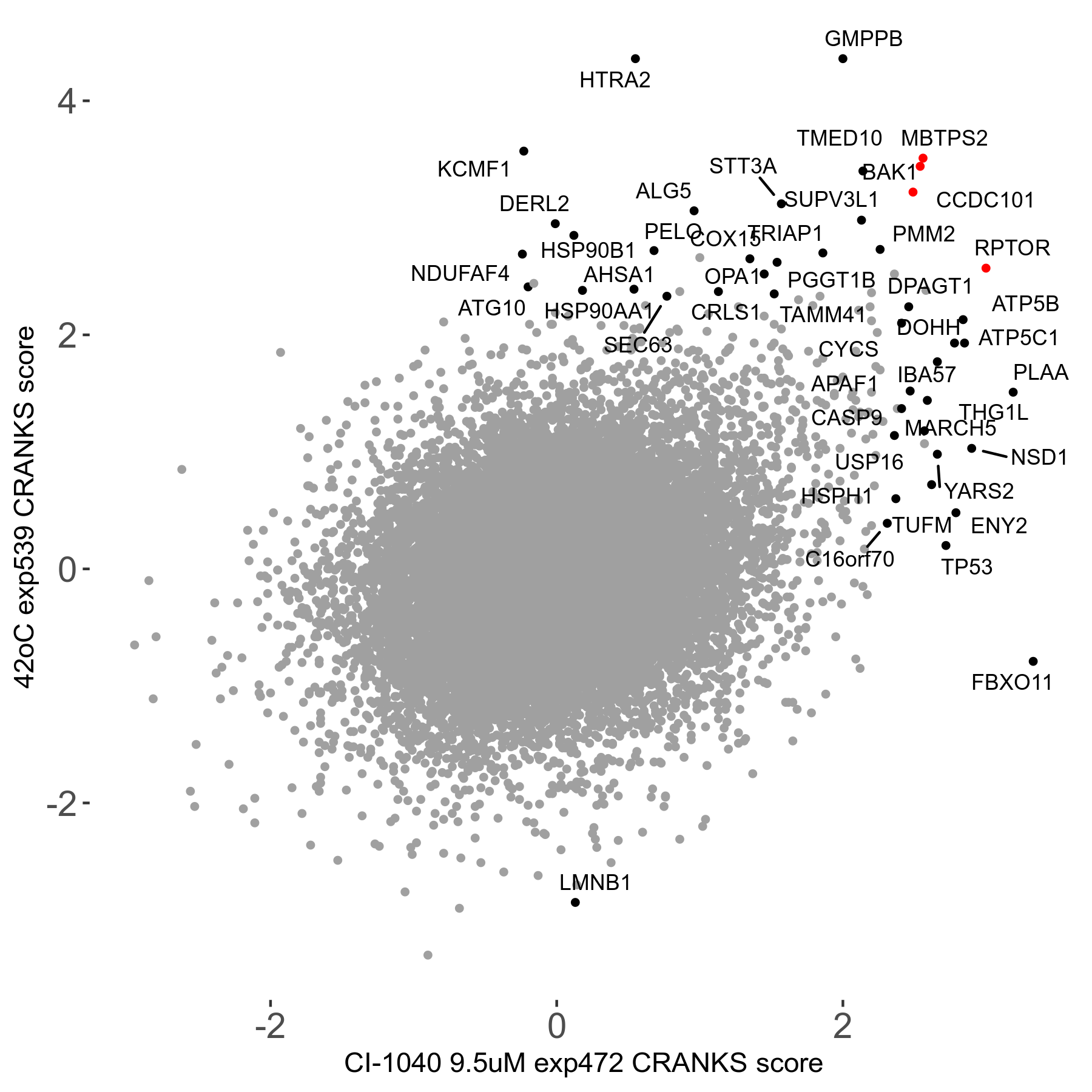
42°C R08 exp539 vs CI-1040 9.5μM R08 exp472
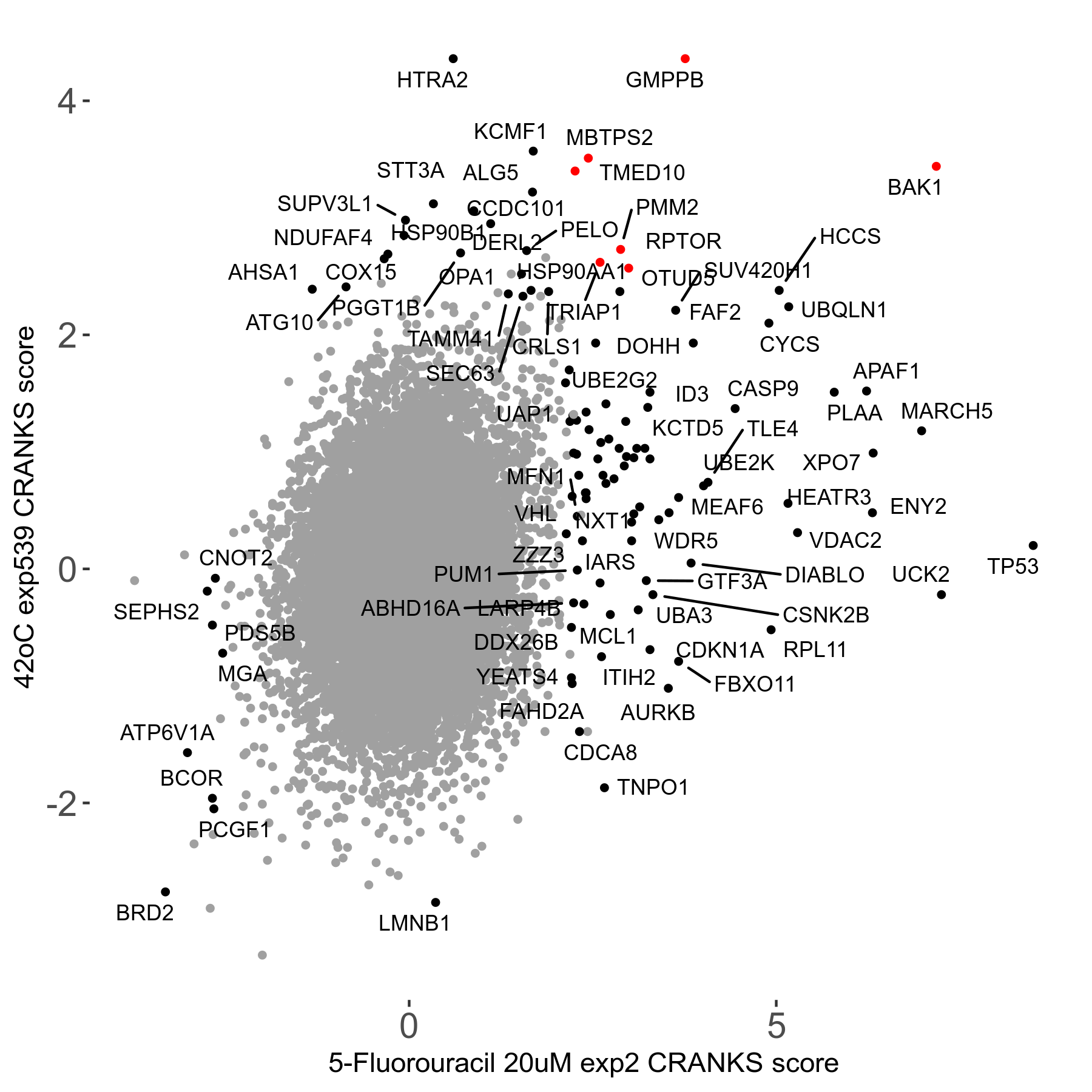
42°C R08 exp539 vs 5-Fluorouracil 20μM R00 exp2
42°C R08 exp539 vs GSK461364A 0.1μM R03 exp126
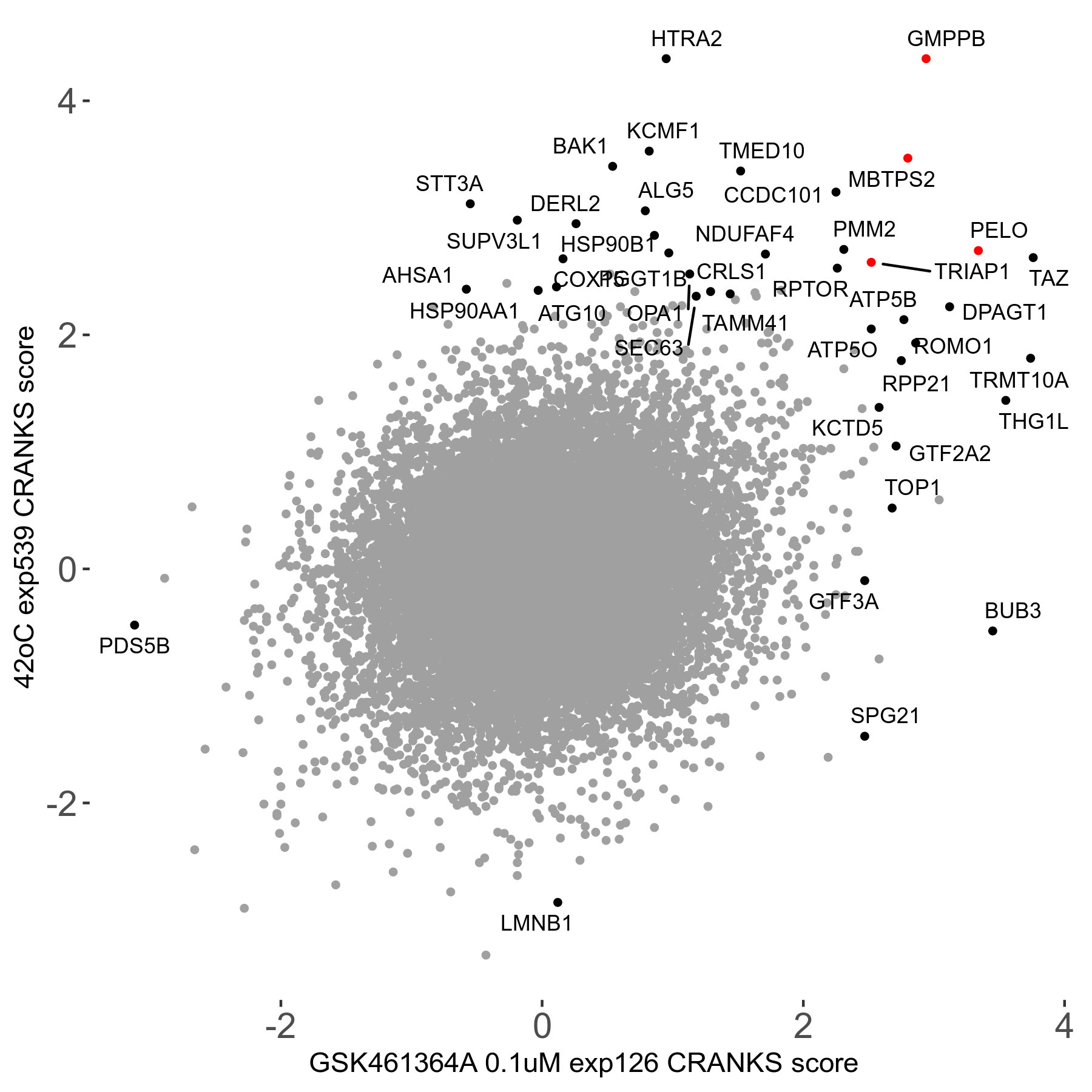
42°C R08 exp539 vs Ammonium tetrathiomolybdate 10μM R08 exp448
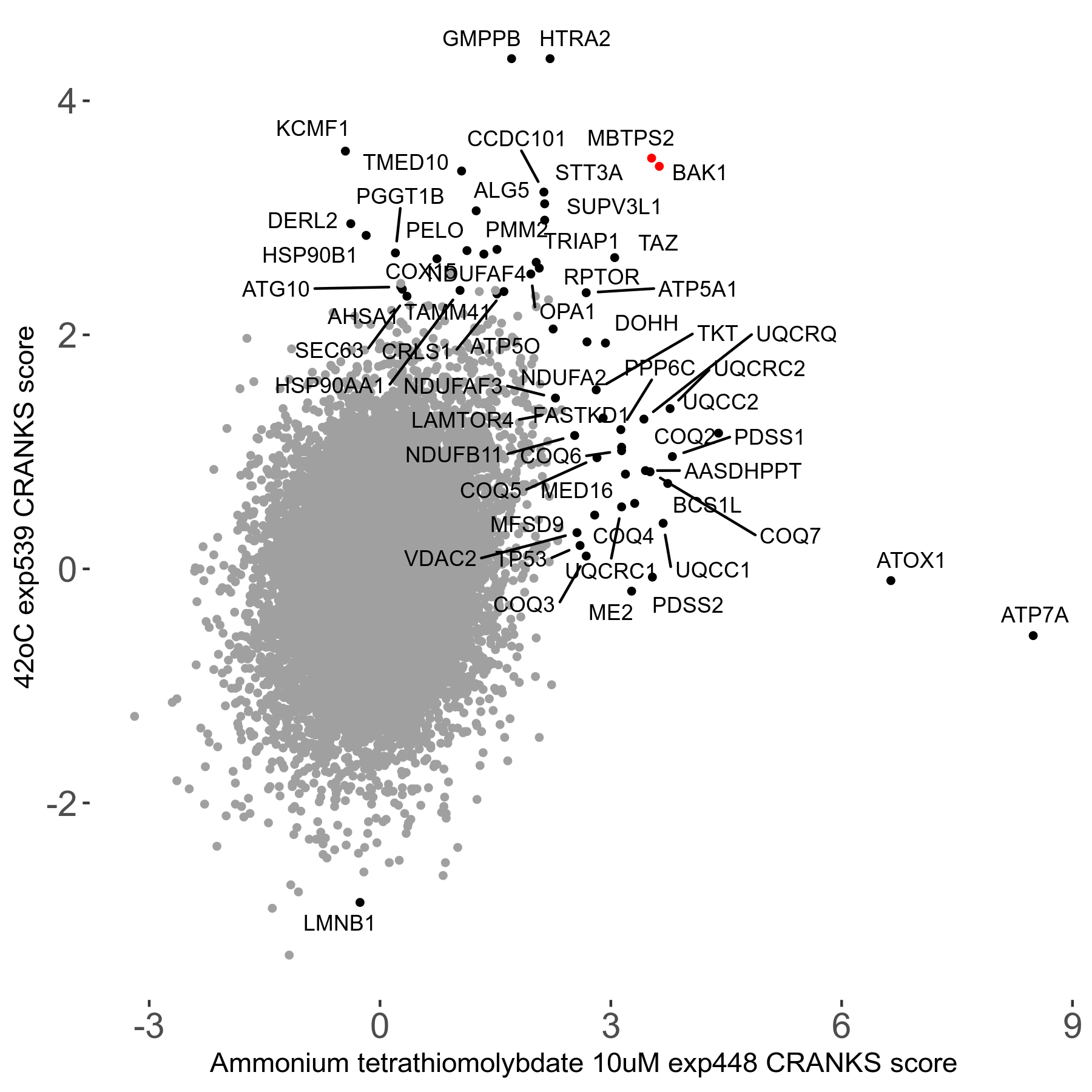
42°C R08 exp539 vs Pimelic-diphenylamide-106 5μM R00 exp28
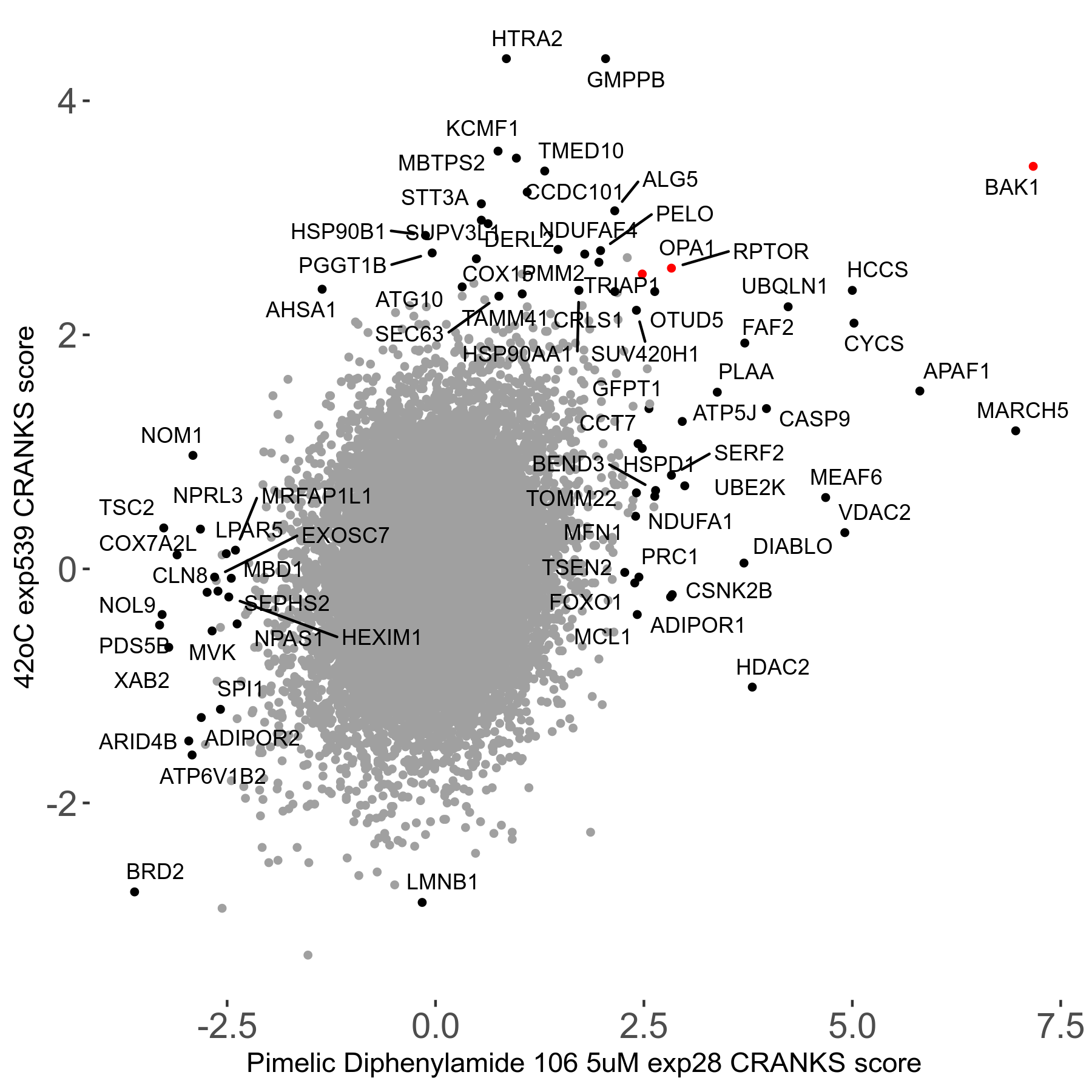
42°C R08 exp539 vs Etoposide 1μM R00 exp19
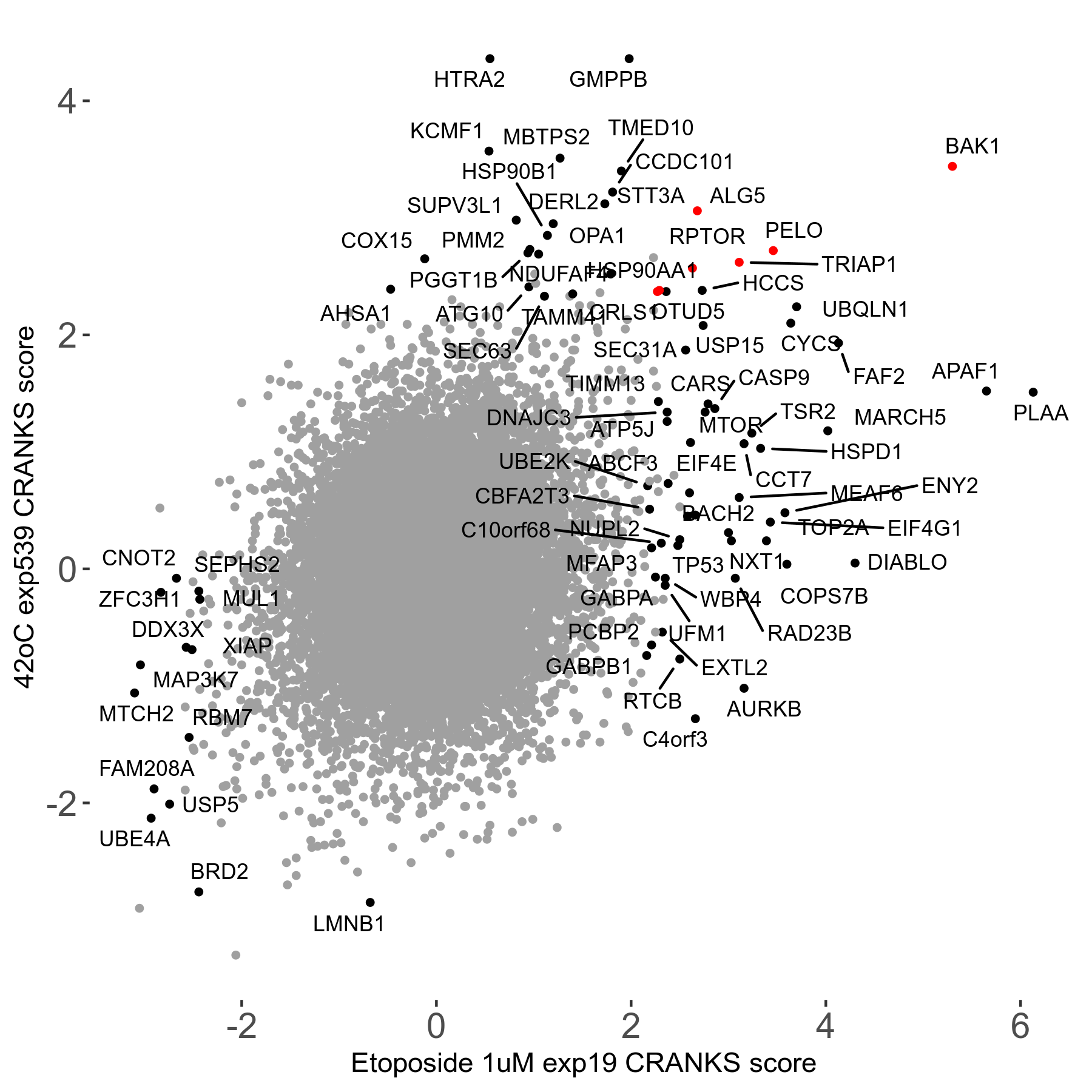
42°C R08 exp539 vs Doxorubicin 0.02μM R08 exp478
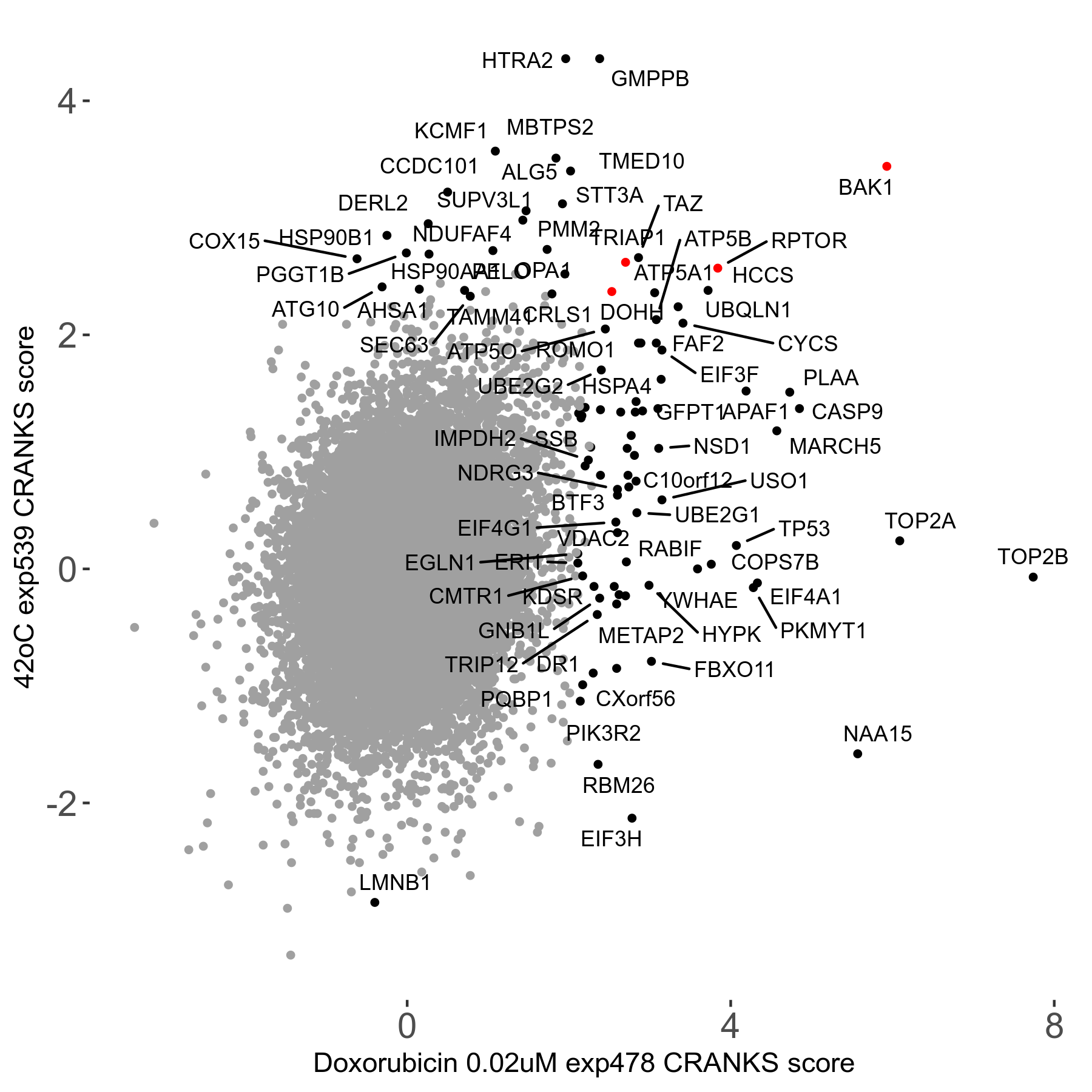
42°C R08 exp539 vs Hippuristanol 0.12μM R08 exp488
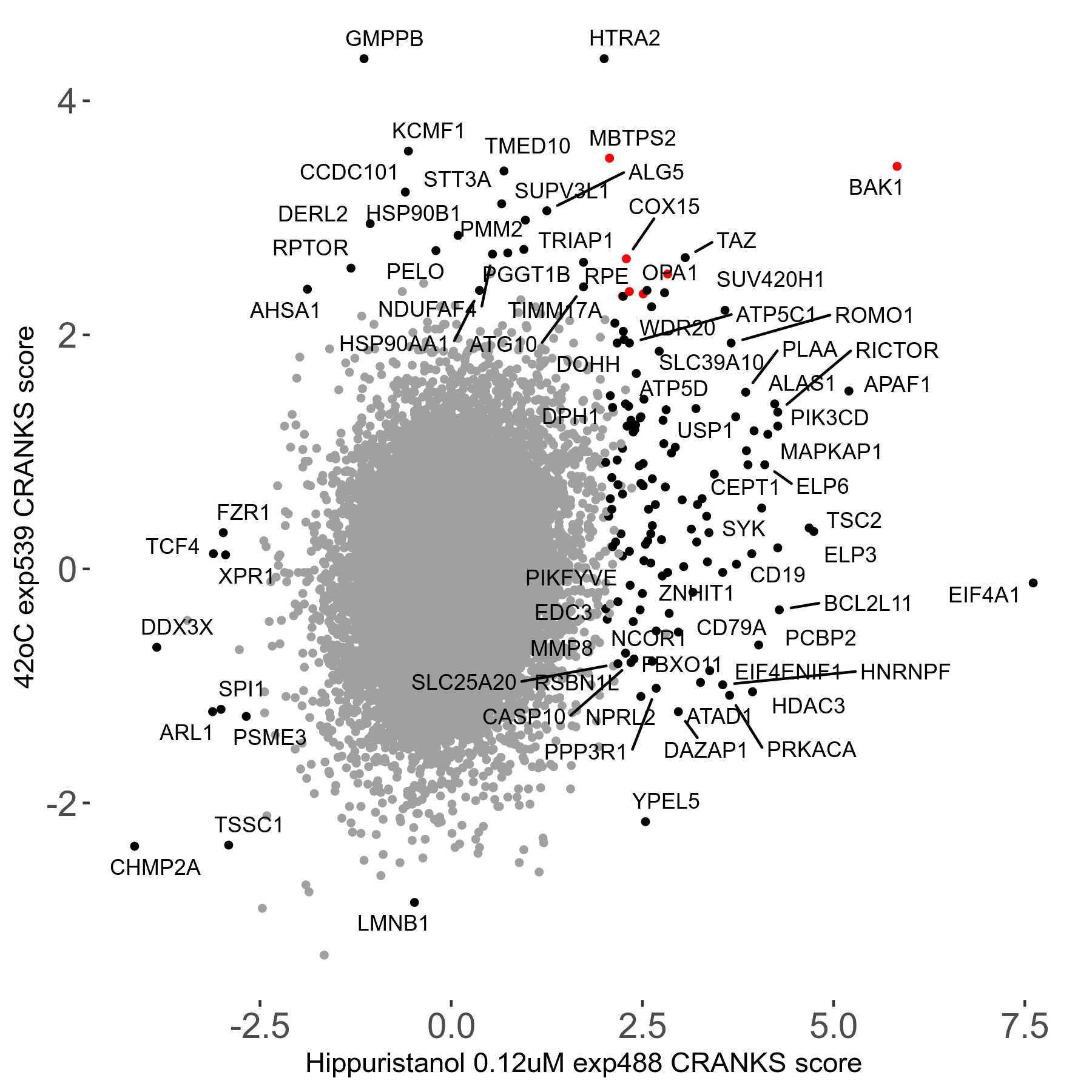
42°C R08 exp539 vs Actinomycin-D 0.001μM R00 exp3
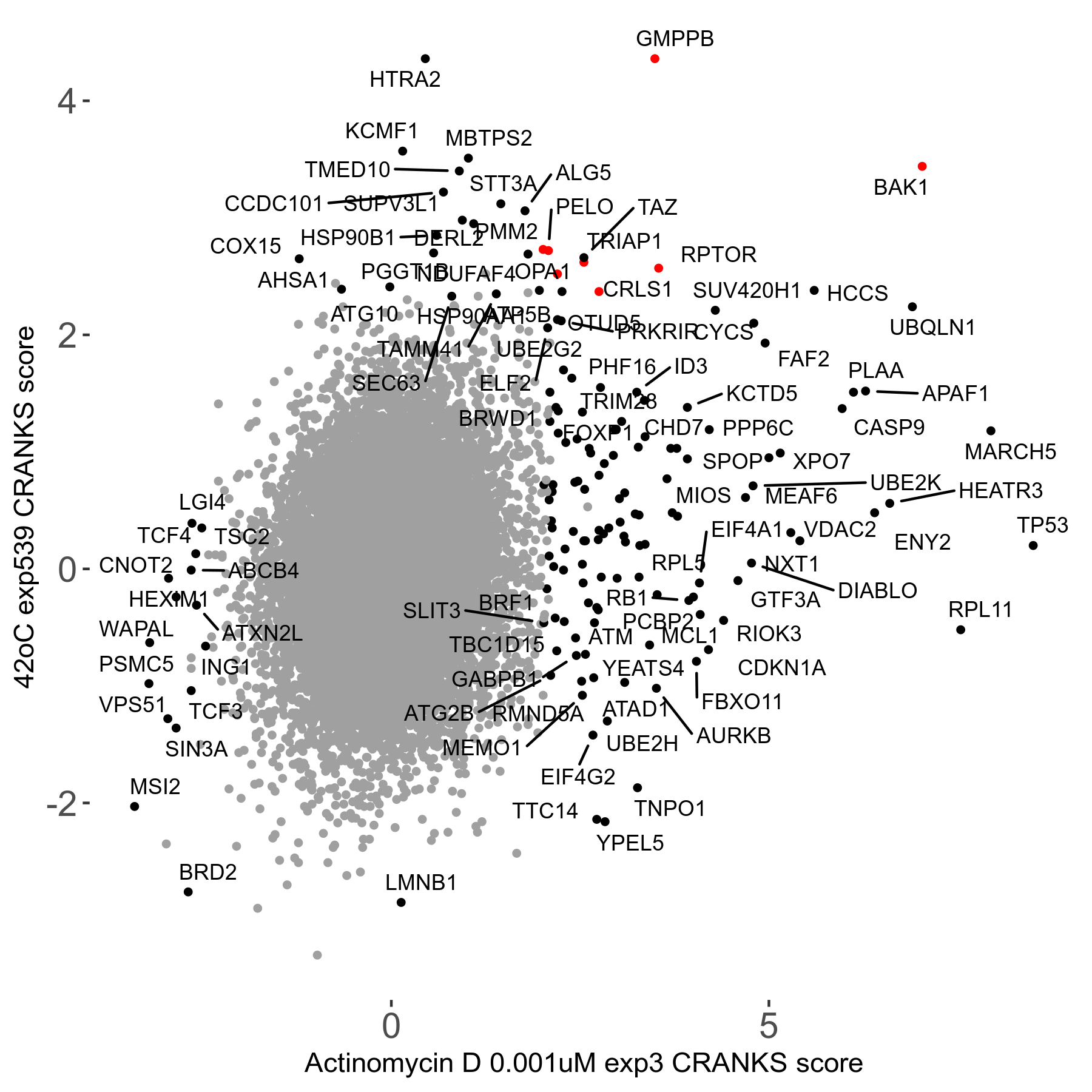
42°C R08 exp539 vs Bleomycin 5μM R08 exp459
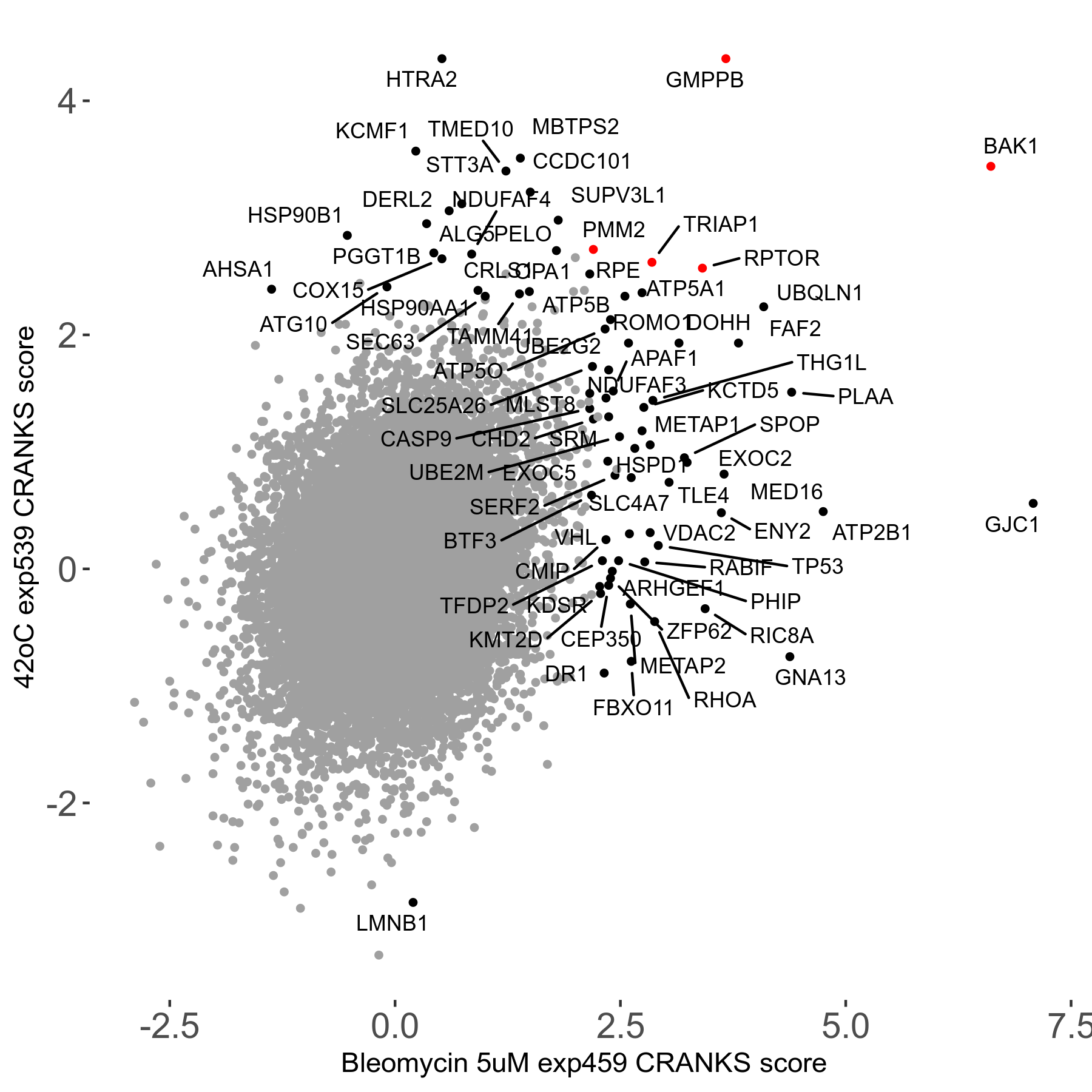
42°C R08 exp539 vs Vinblastine 0.2μM R03 exp108
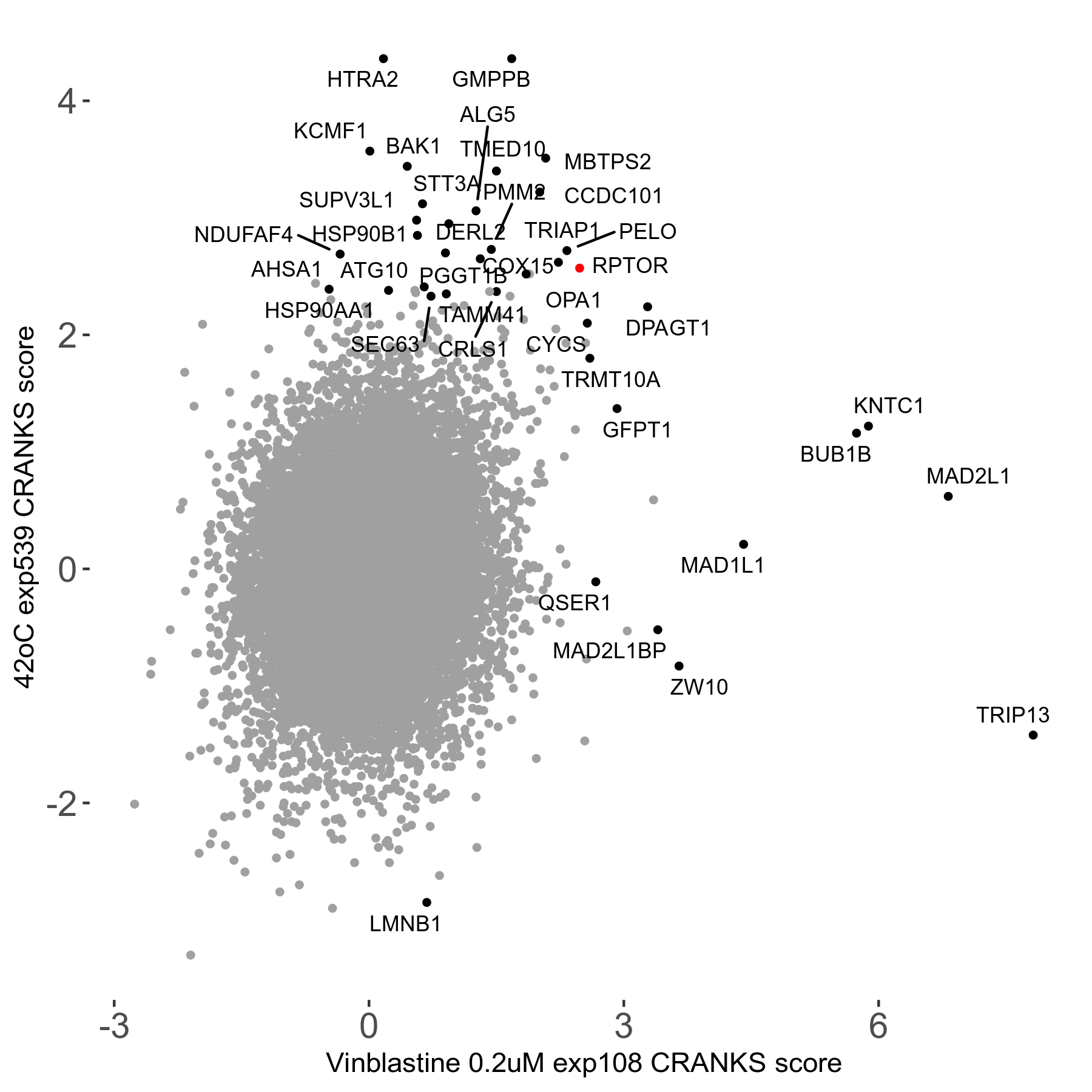
42°C R08 exp539 vs Tanespimycin 14μM R08 exp527
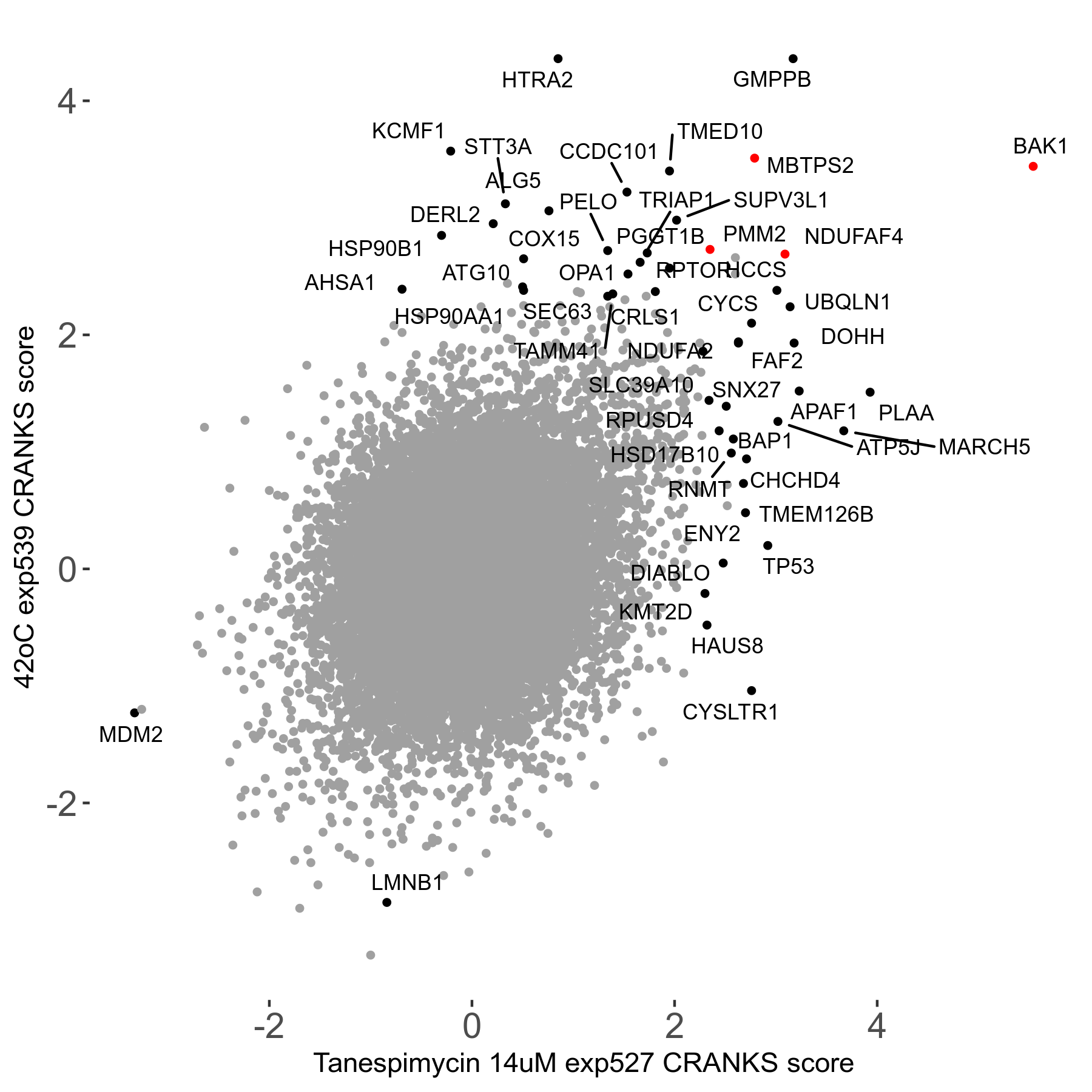
42°C R08 exp539 vs Atovaquone 15μM R08 exp451
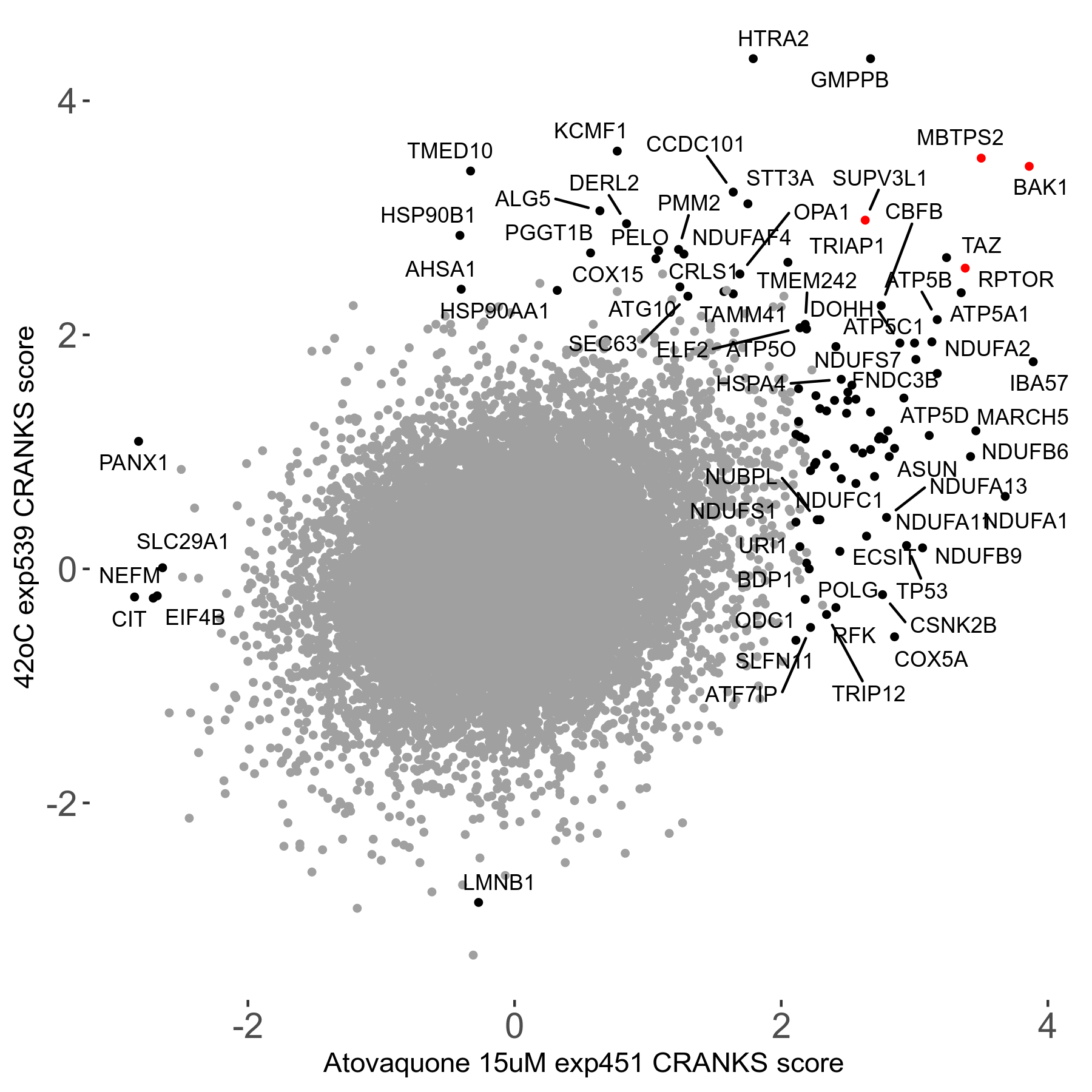
42°C R08 exp539 vs Benzoate 20000μM R08 exp456
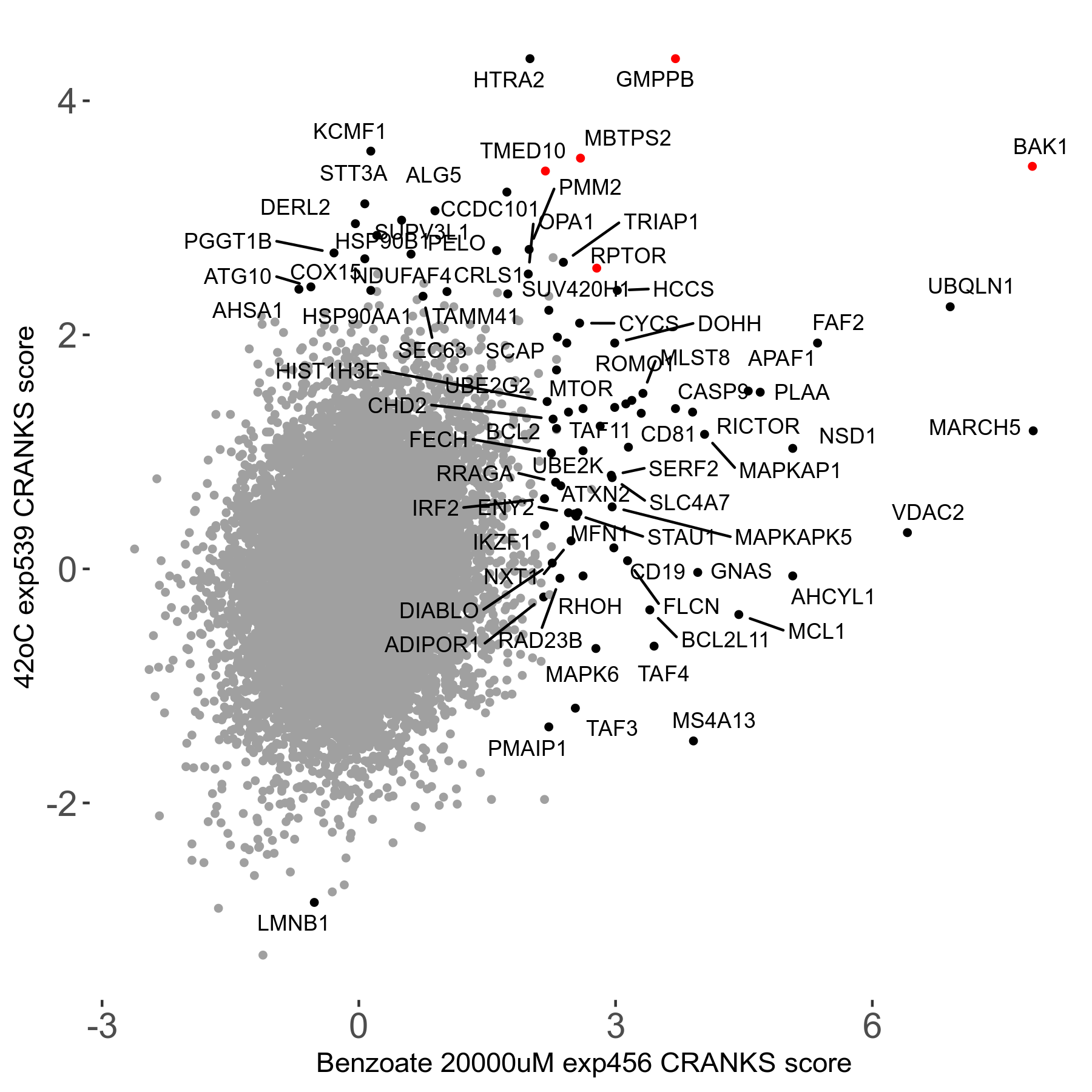
42°C R08 exp539 vs Aphidicolin 0.4μM R08 exp440
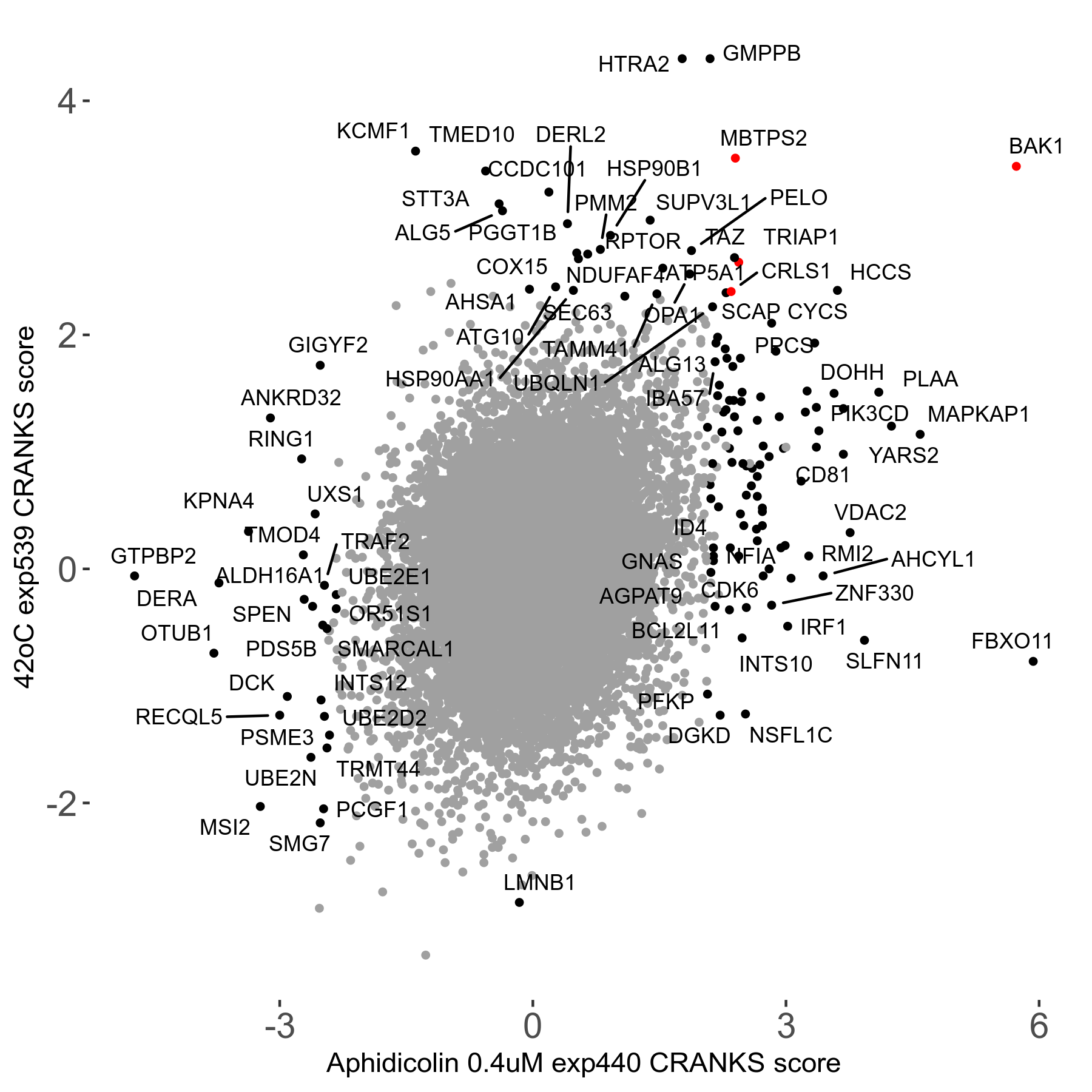
42°C R08 exp539 vs MLN-4924 2μM R00 exp22
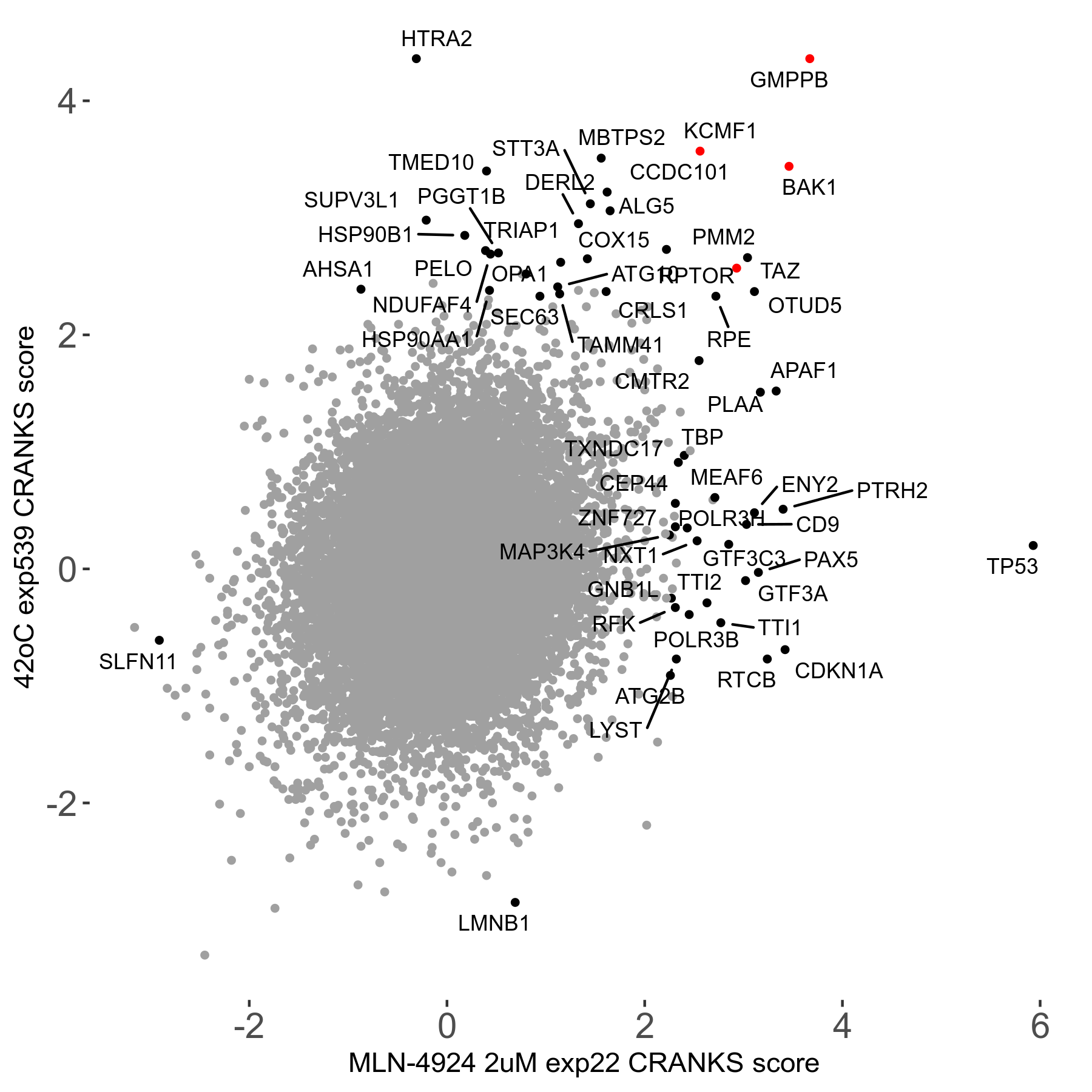
42°C R08 exp539 vs BI-2536 0.02μM R03 exp96
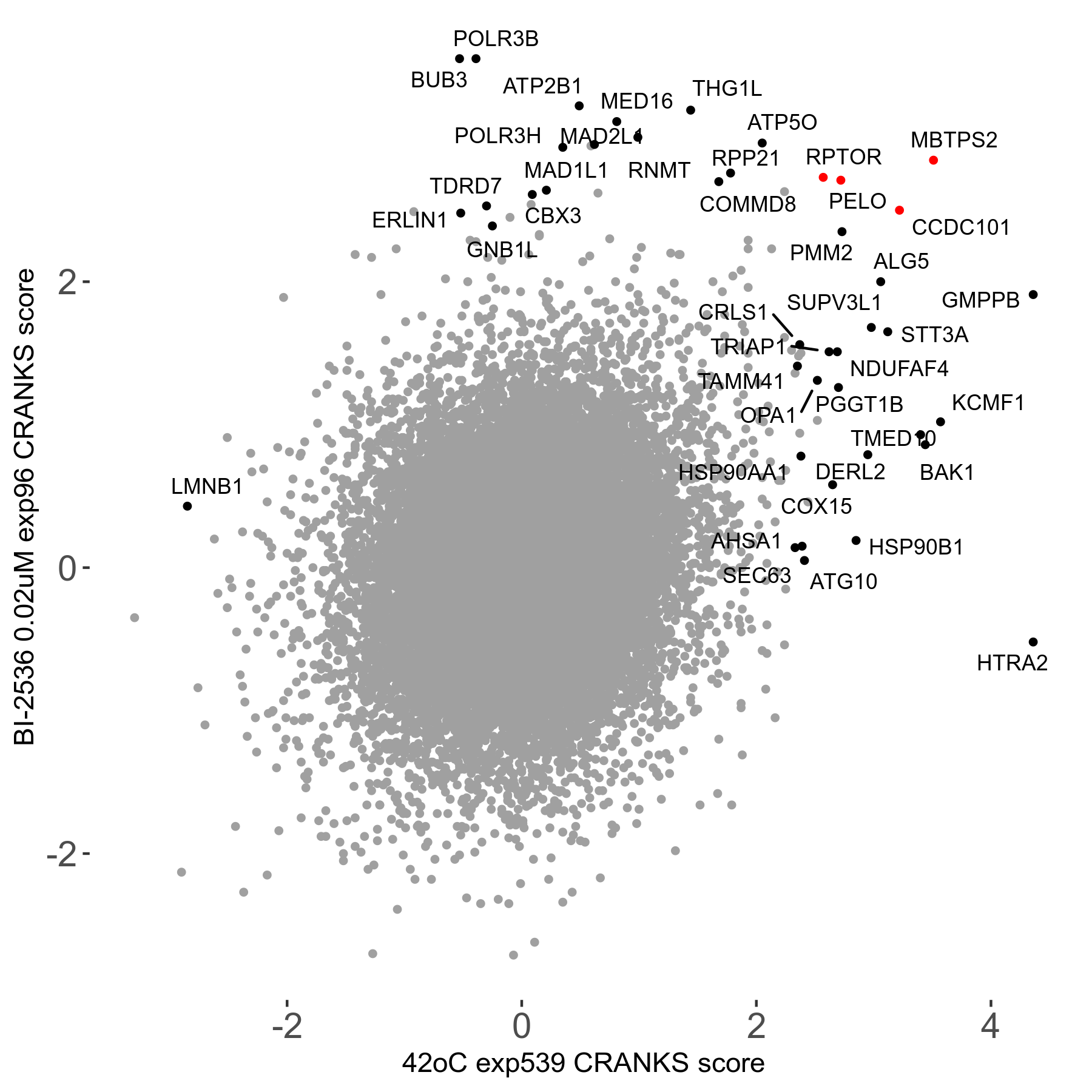
42°C R08 exp539 vs UM131593 0.2μM R03 exp106
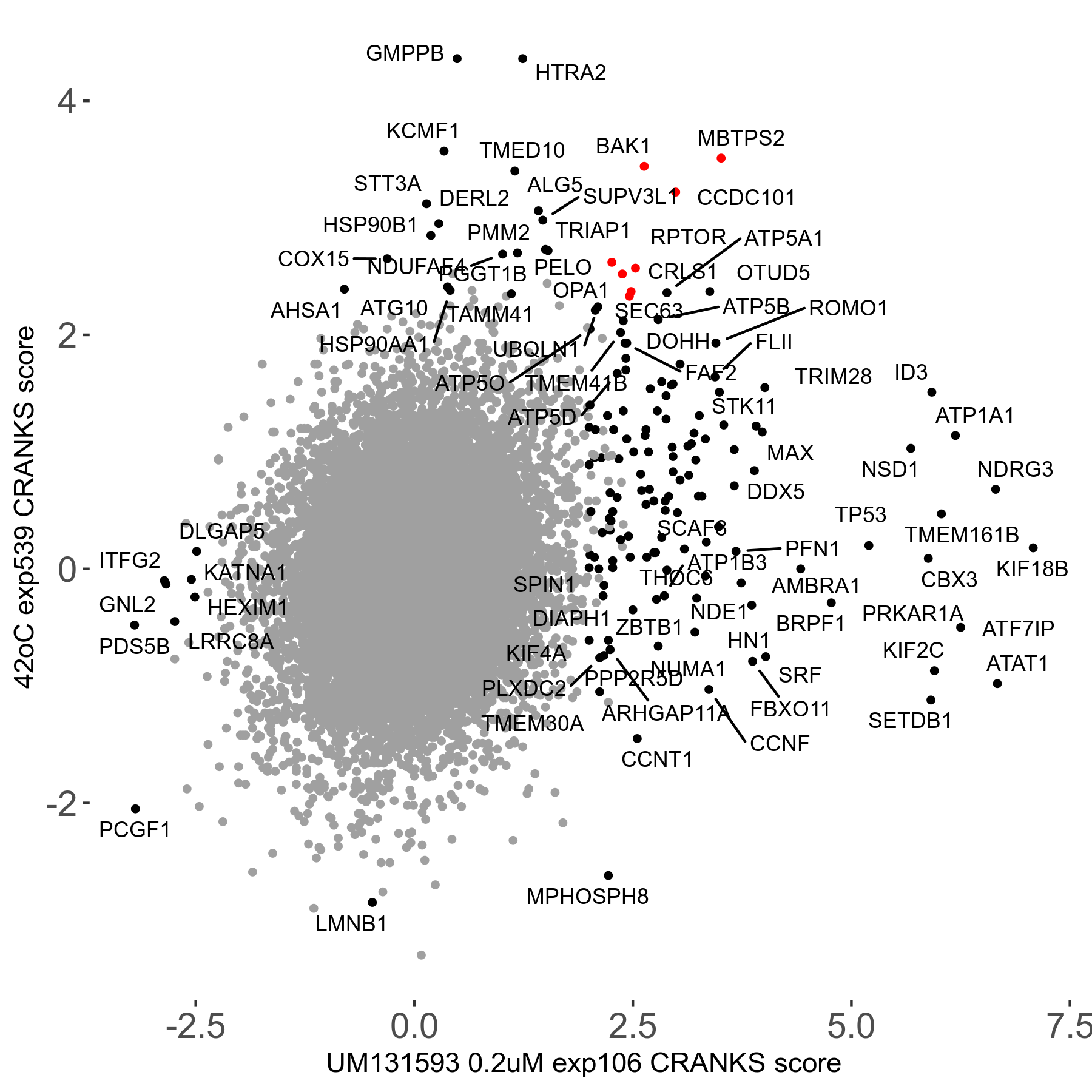
42°C R08 exp539 vs Mitomycin-C 0.06μM R08 exp503
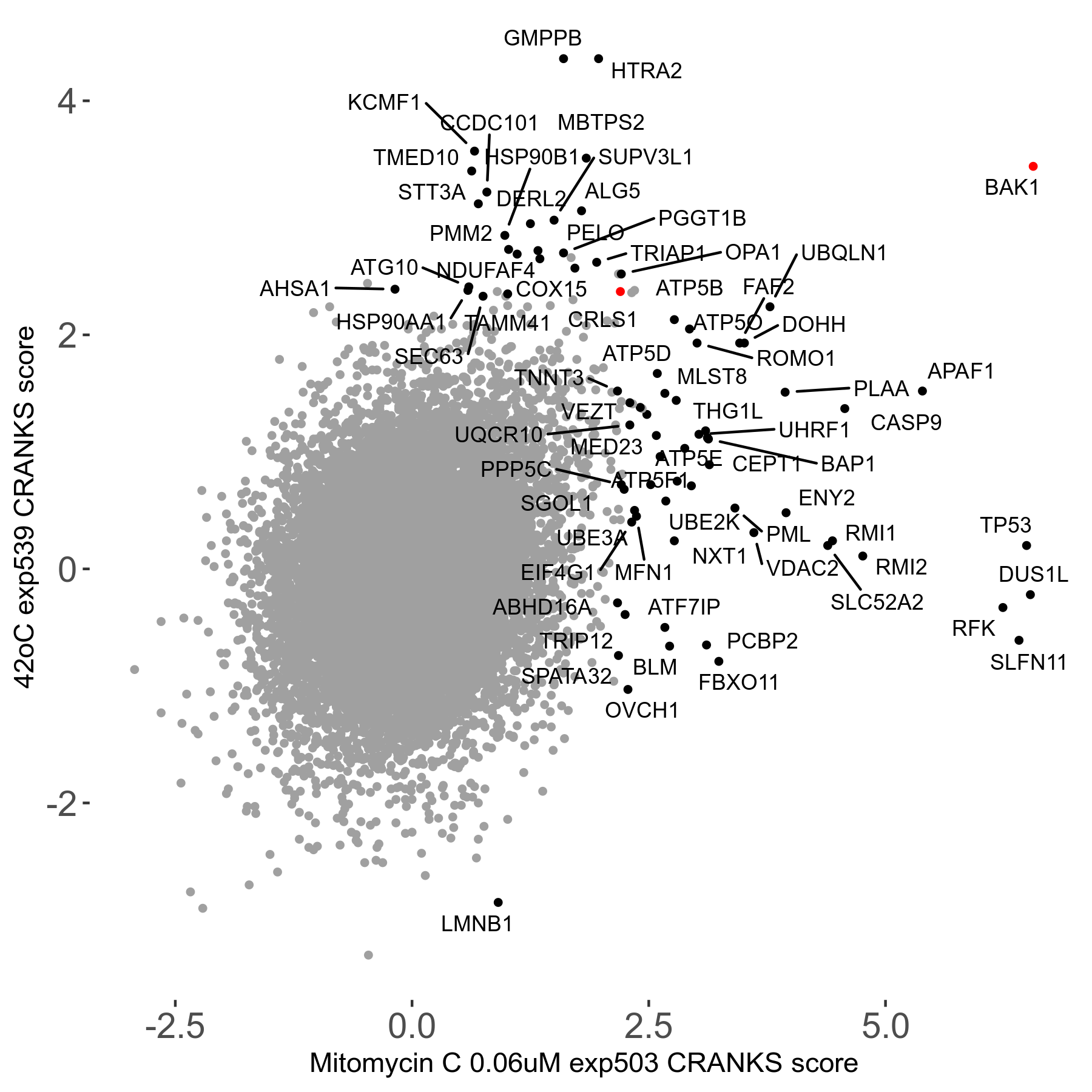
42°C R08 exp539 vs THZ1 0.06μM R08 exp531
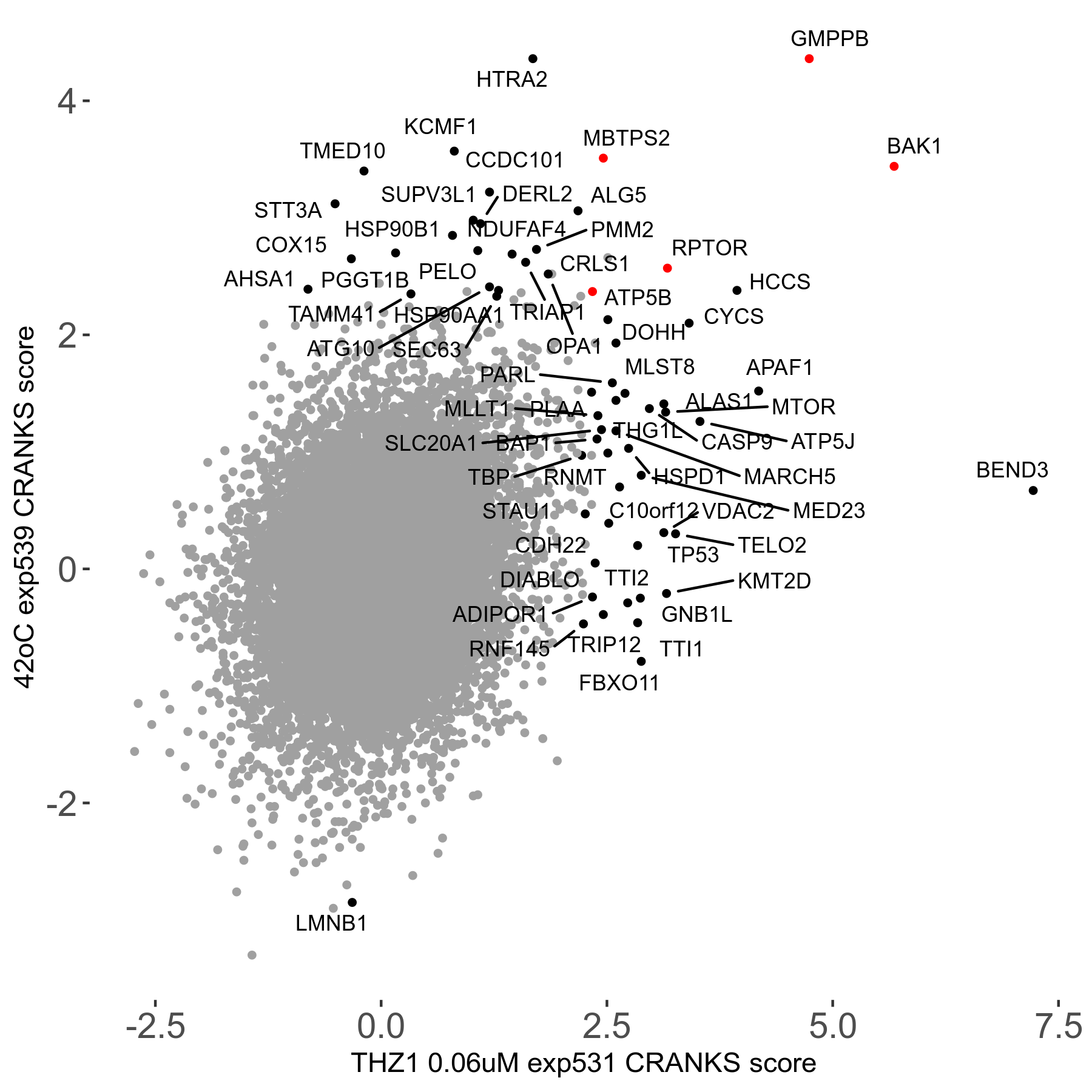
42°C R08 exp539 vs Hinokiflavone 12μM R08 exp487
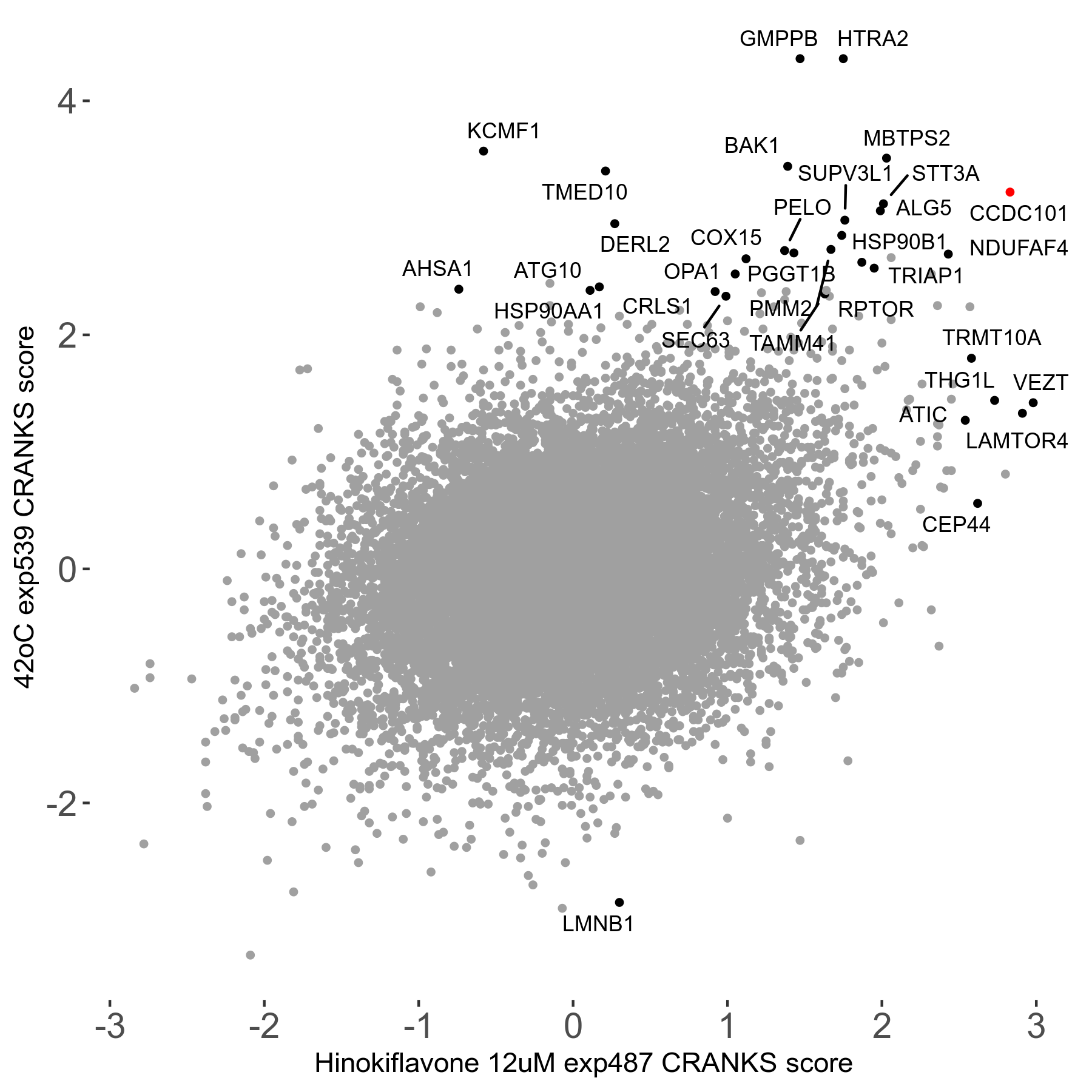
42°C R08 exp539 vs Hippuristanol 0.12μM R08 exp489 no dilution day6
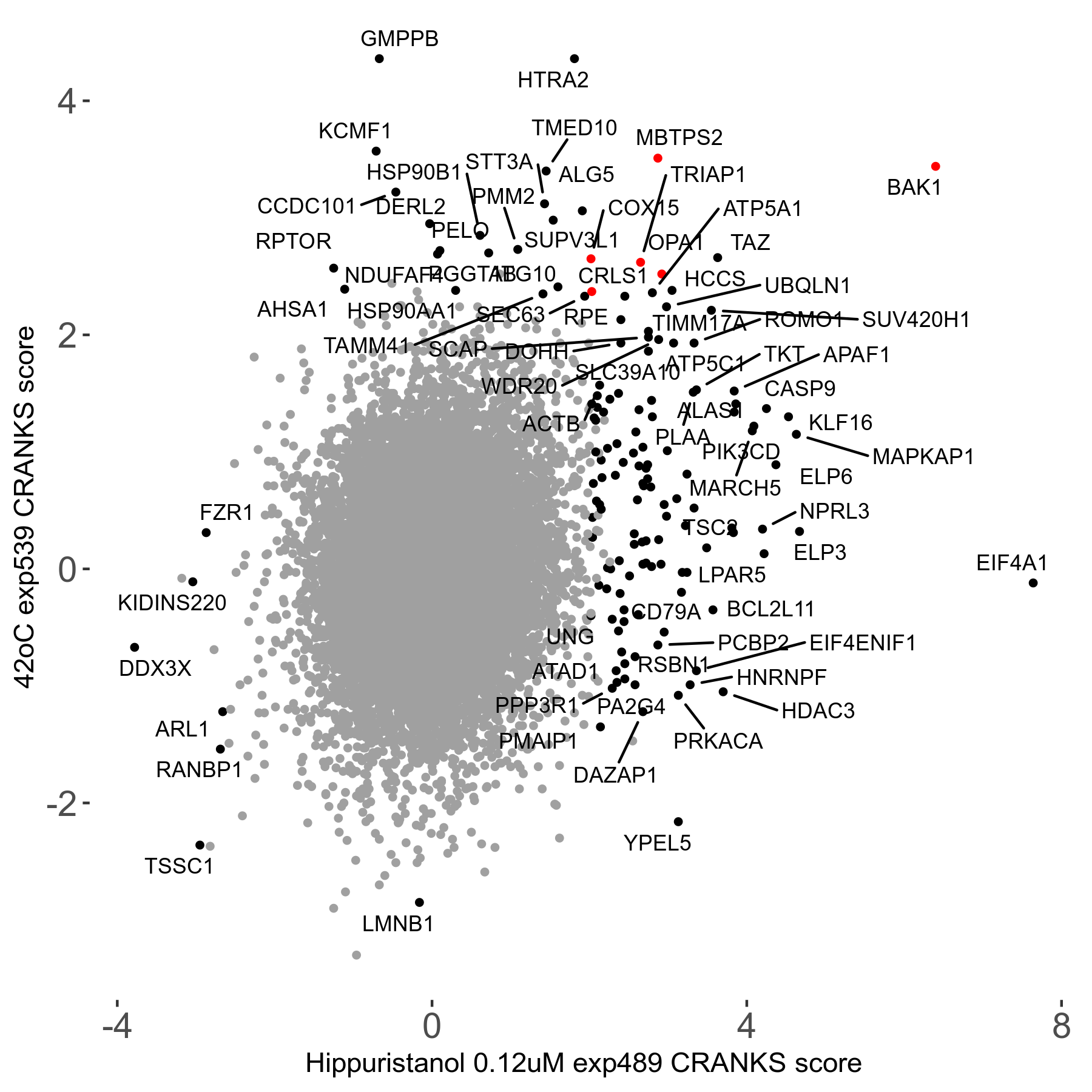
42°C R08 exp539 vs Thimerosal 0.85μM R08 exp529
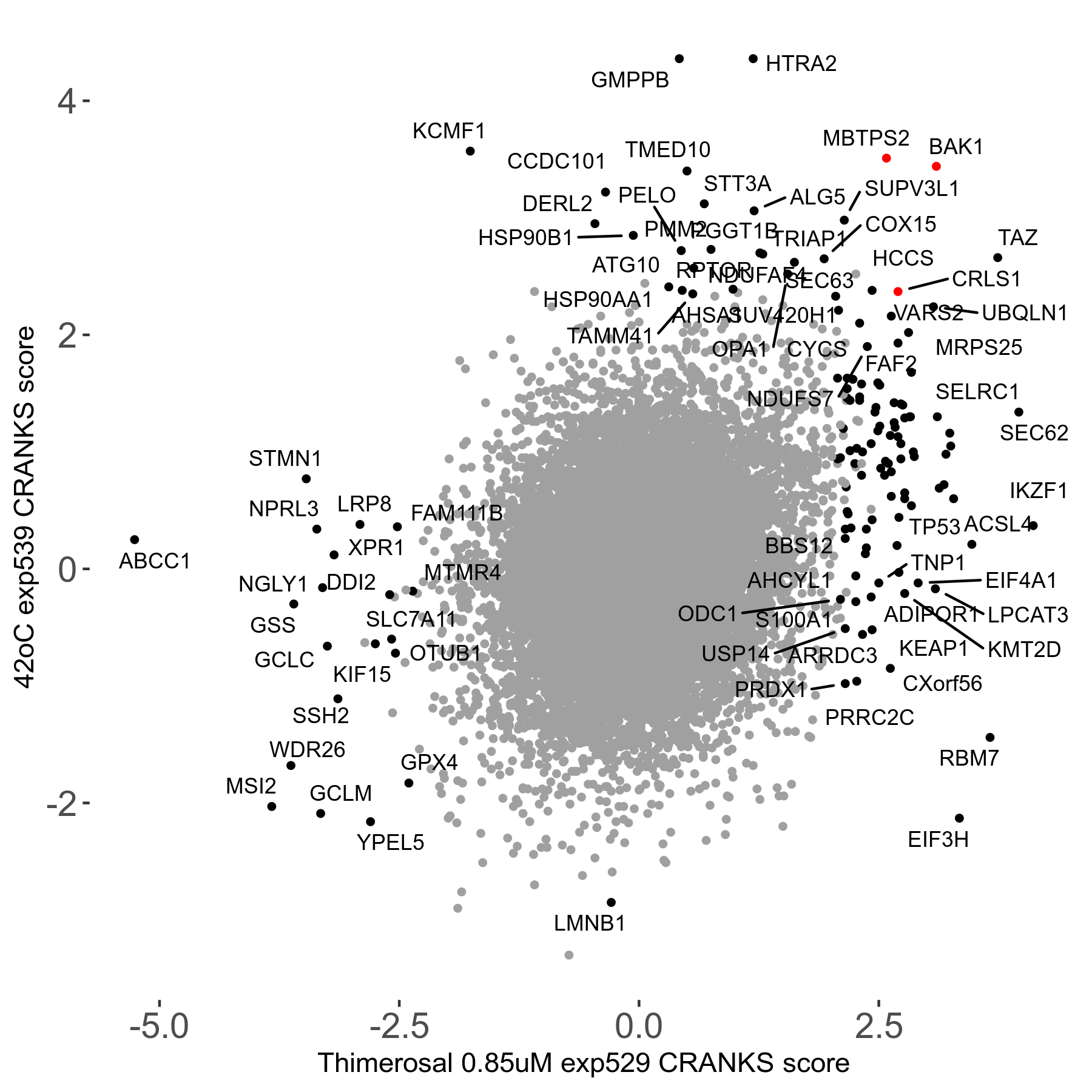
42°C R08 exp539 vs CFI-400945 25μM R08 exp469
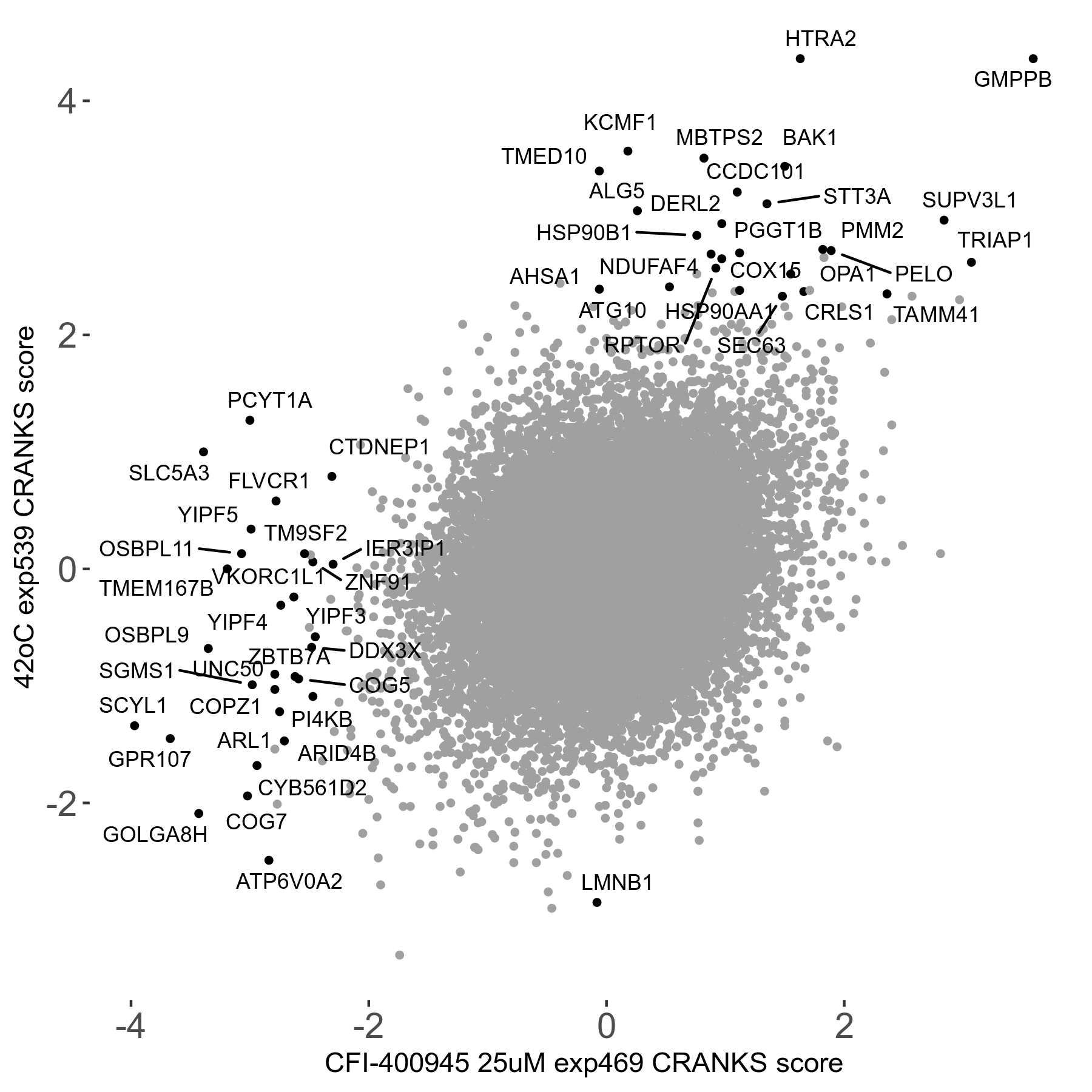
42°C R08 exp539 vs Etoposide 0.3μM R05 exp199
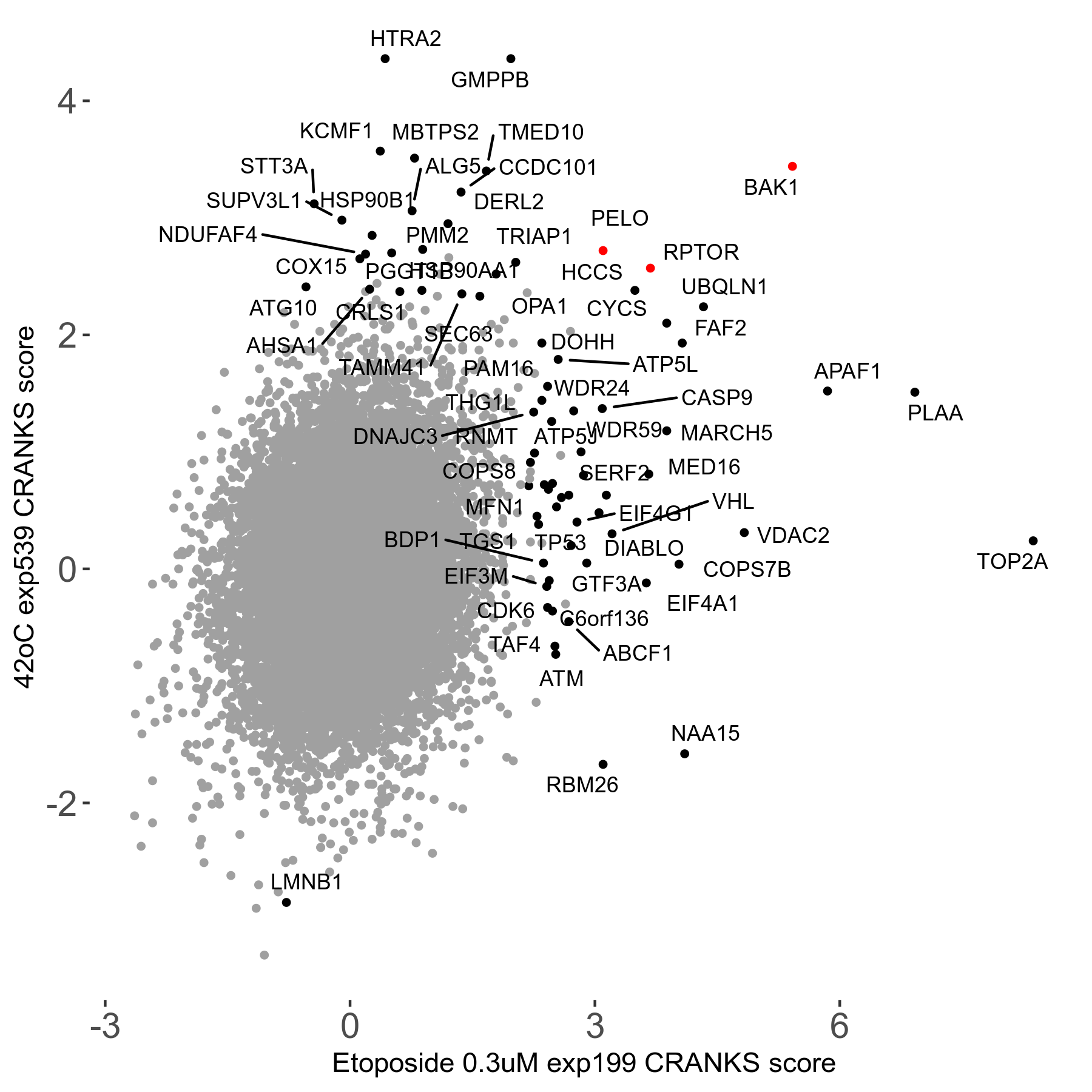
42°C R08 exp539 vs Rotenone 20μM R00 exp34
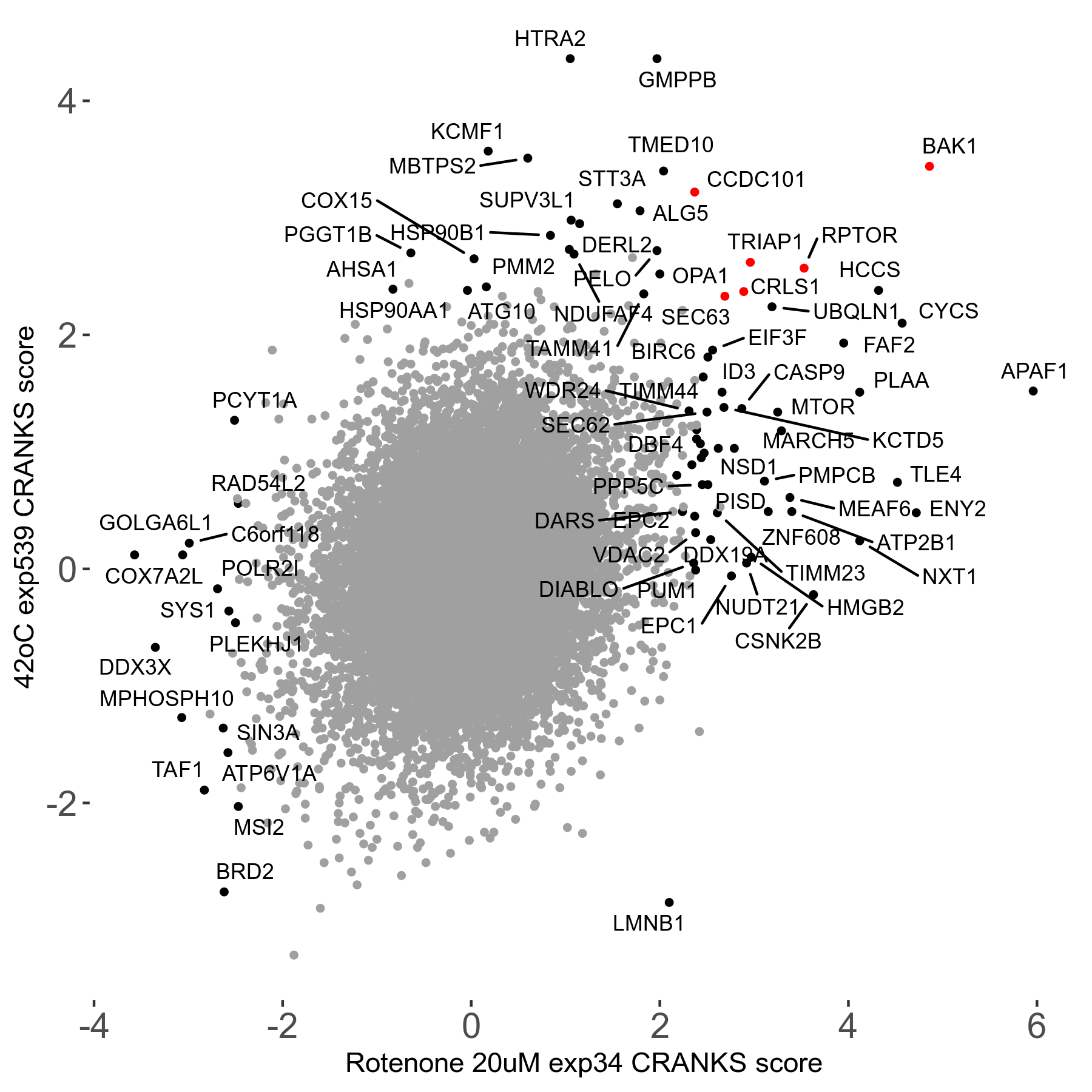
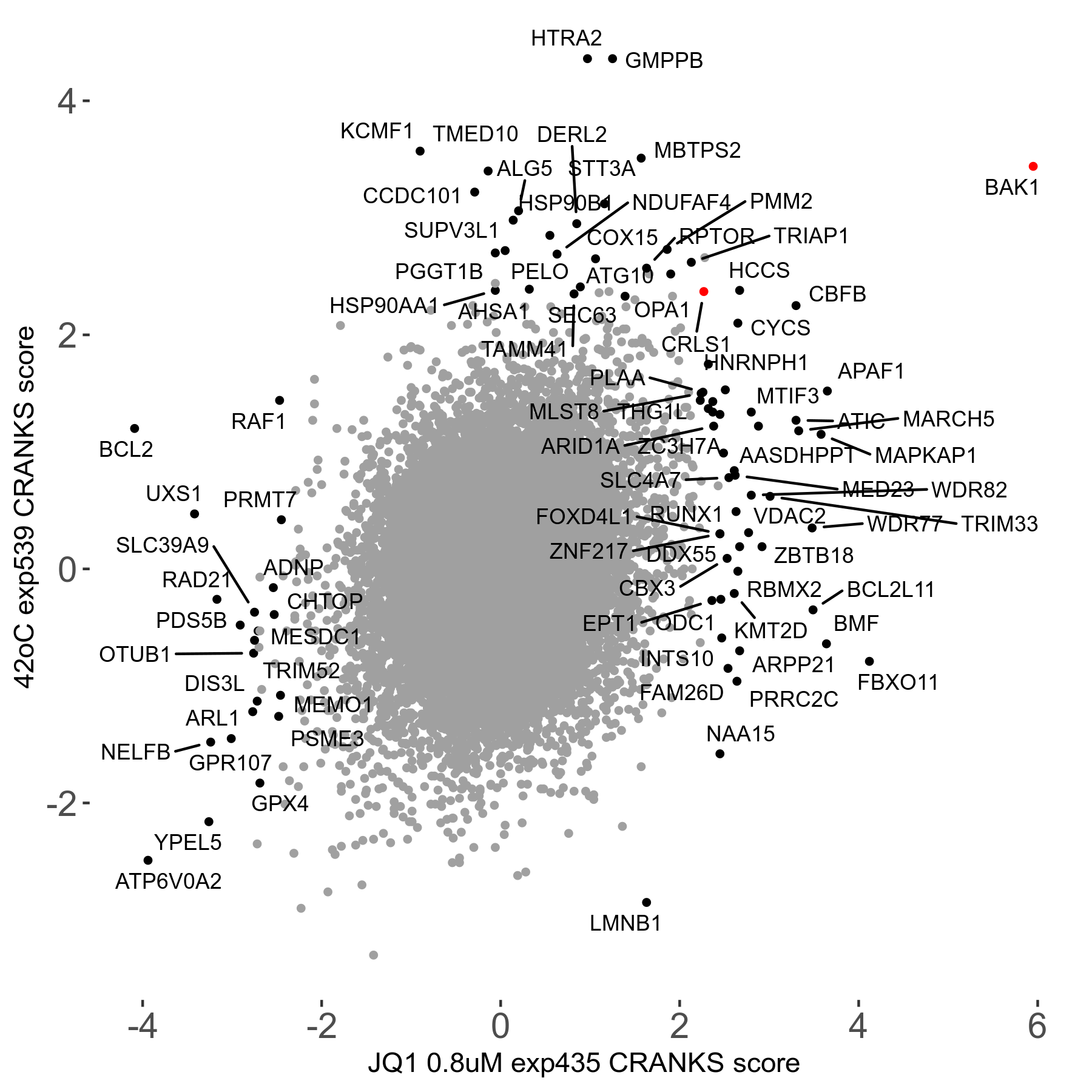
42°C R08 exp539 vs JQ1 0.8μM R08 exp435
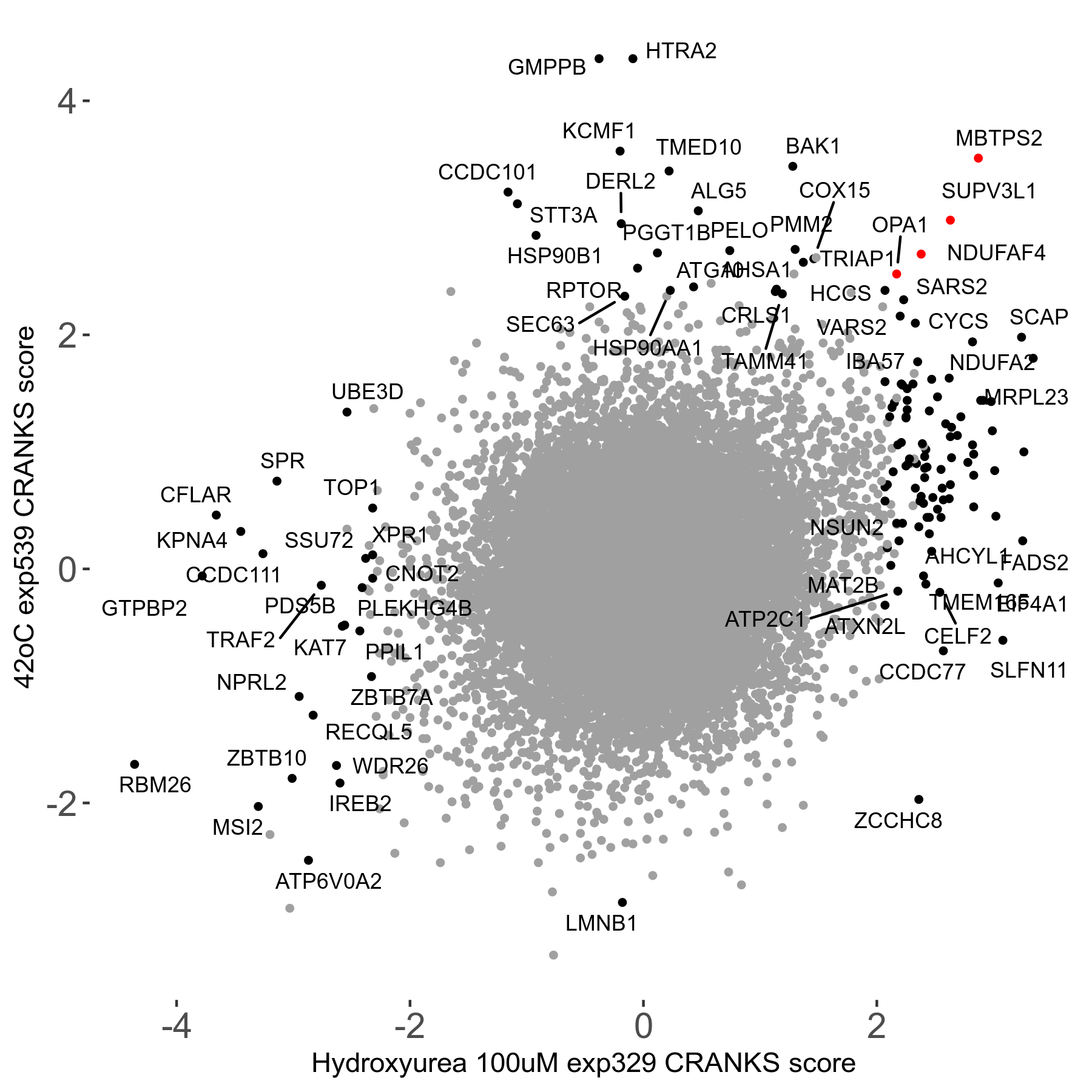
42°C R08 exp539 vs Hydroxyurea 100μM R07 exp329
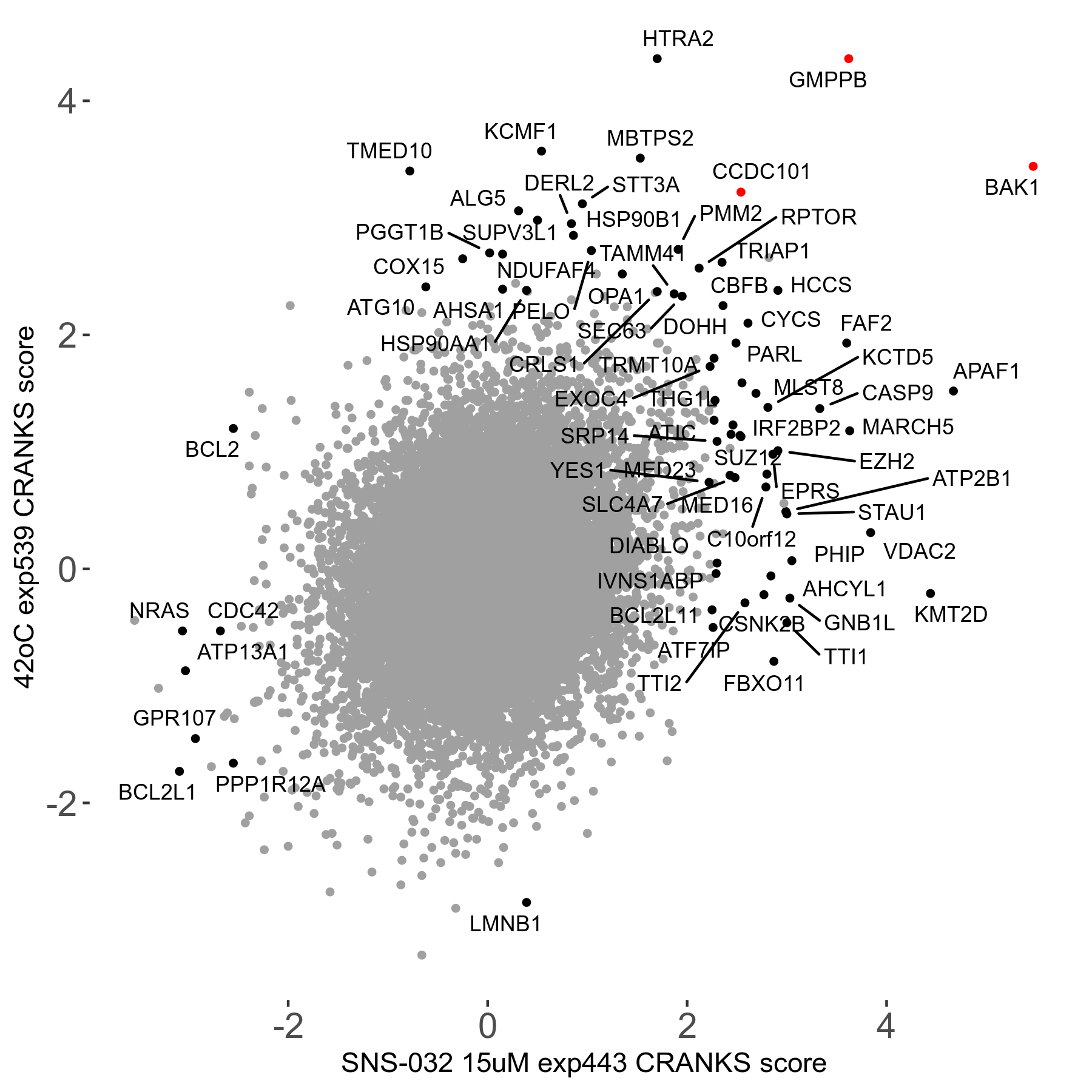
42°C R08 exp539 vs SNS-032 15μM R08 exp443
Top 30 sensitive GO terms
| GO Term | Fold Change | Genes |
|---|---|---|
| vesicle tethering complex | 63.48 | VPS52,COG1,RINT1,COG3,TRAPPC13,TRAPPC2L |
| spliceosomal complex assembly | 32.65 | DDX23,PRPF19,SF3A1,HTATSF1 |
| Golgi vesicle transport | 16.62 | VPS52,NAPA,COG1,CLN3,RINT1,COG3,TRAPPC13,TRAPPC2L |
| endomembrane system organization | 8.66 | VPS25,LMNB1,USP8,COG1,CHMP2A,CLN3,COG3,BROX |
Top 30 resistant GO terms
| GO Term | Fold Change | Genes |
|---|---|---|
| mitochondrial envelope | 7.35 | HTRA2,BAK1,NDUFAF4,COX15,TRIAP1,OPA1,HCCS,CRLS1,ATP5F1A,TAMM41 |
| mitochondrial membrane | 7.05 | HTRA2,BAK1,NDUFAF4,COX15,OPA1,HCCS,CRLS1,ATP5F1A,TAMM41 |