Table of Contents
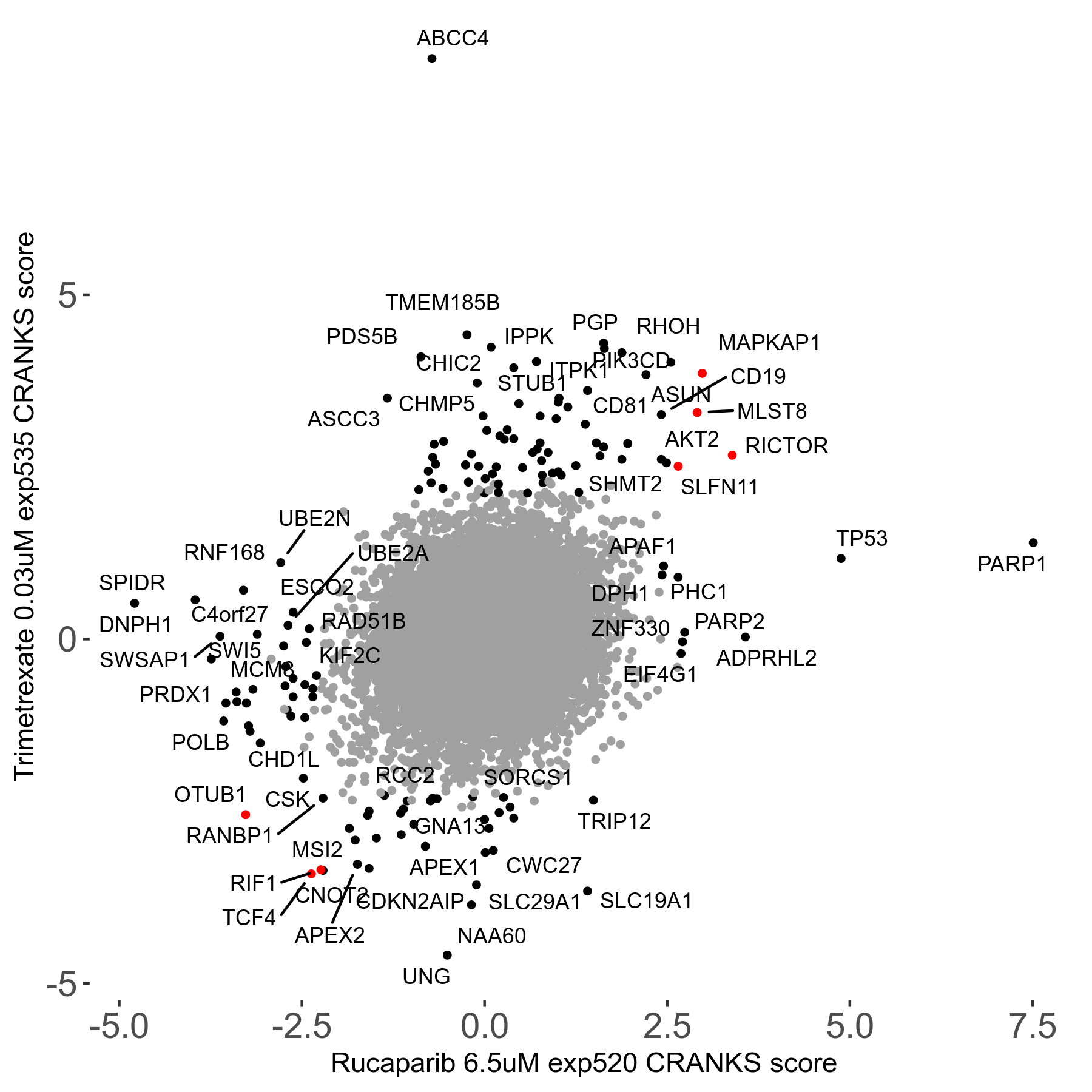
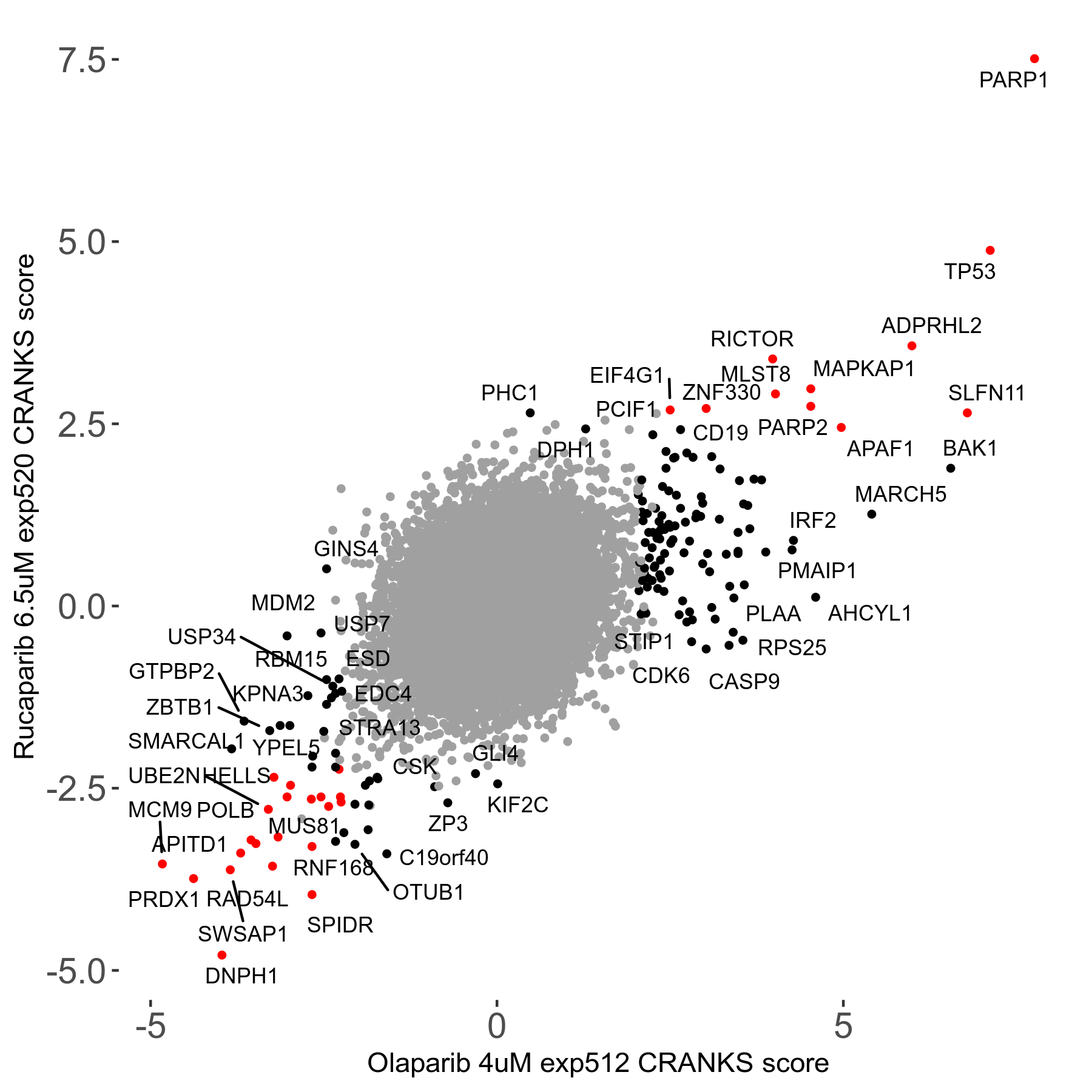
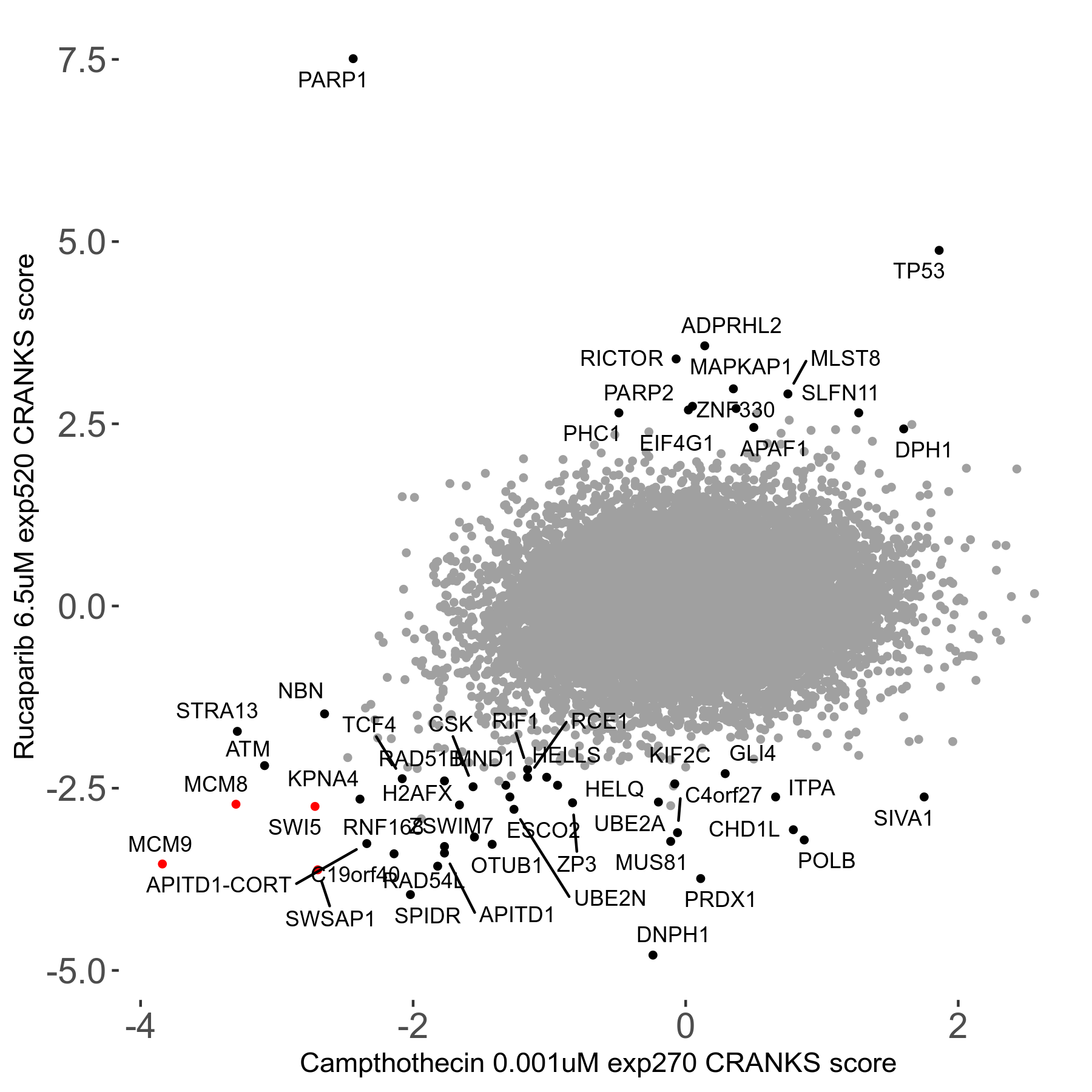
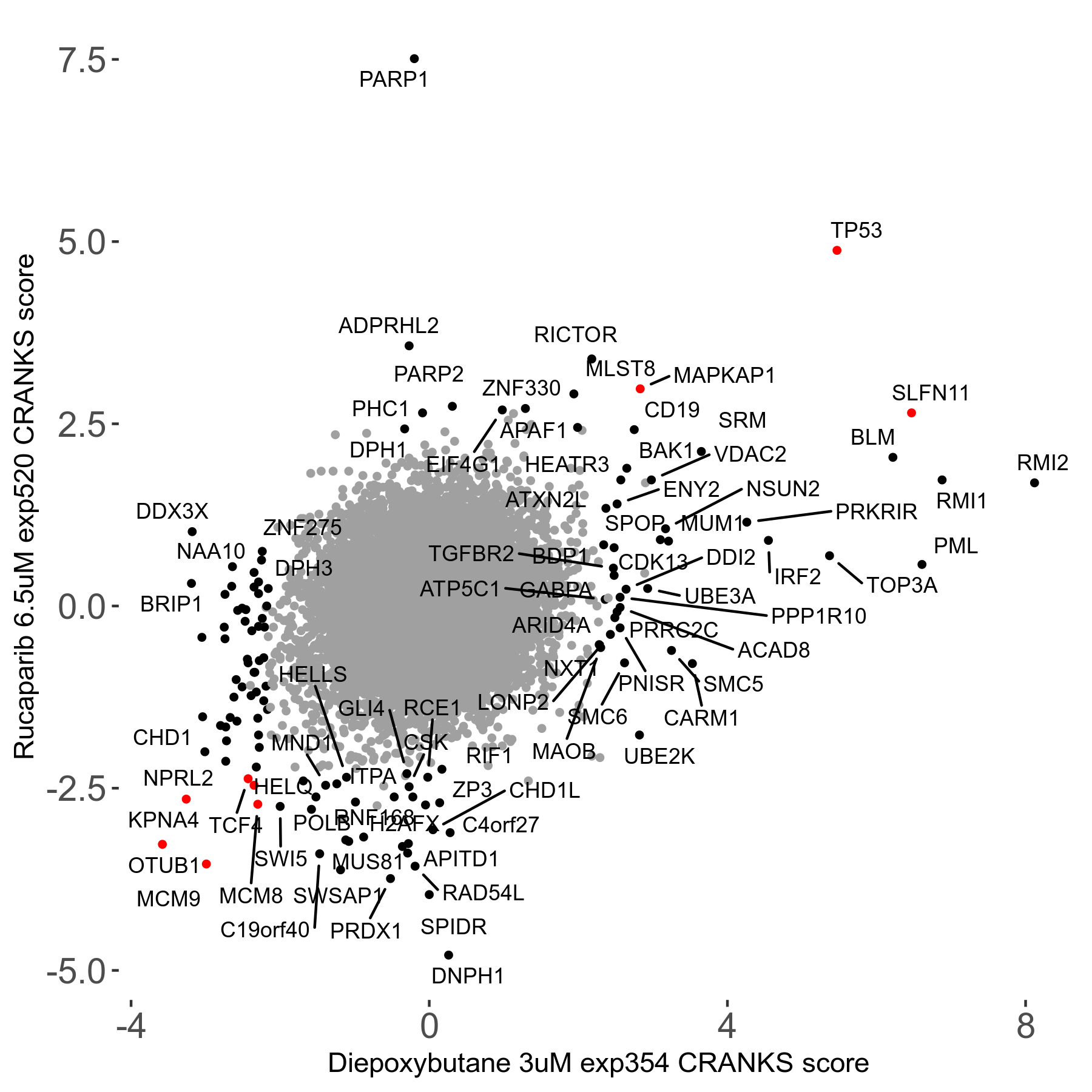
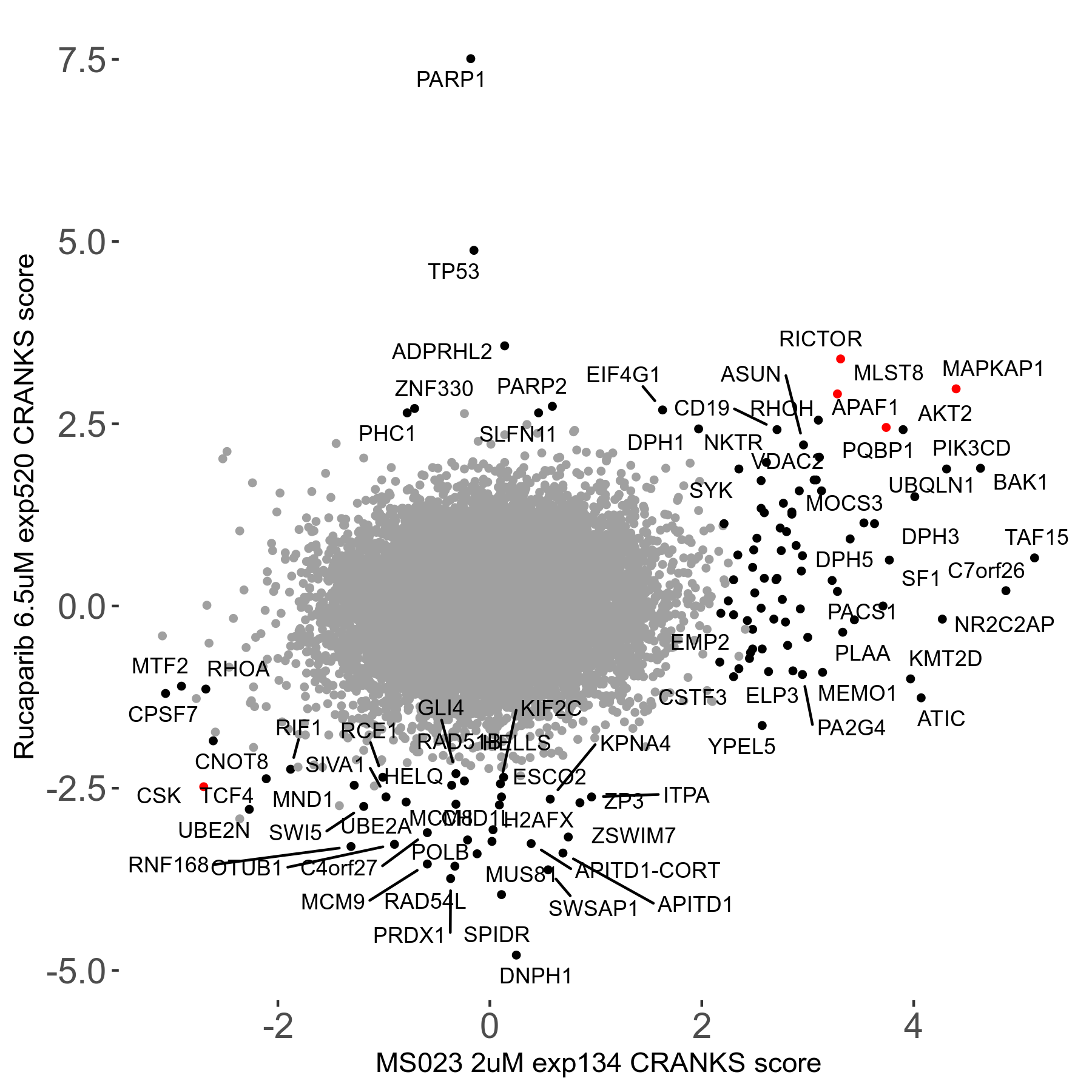
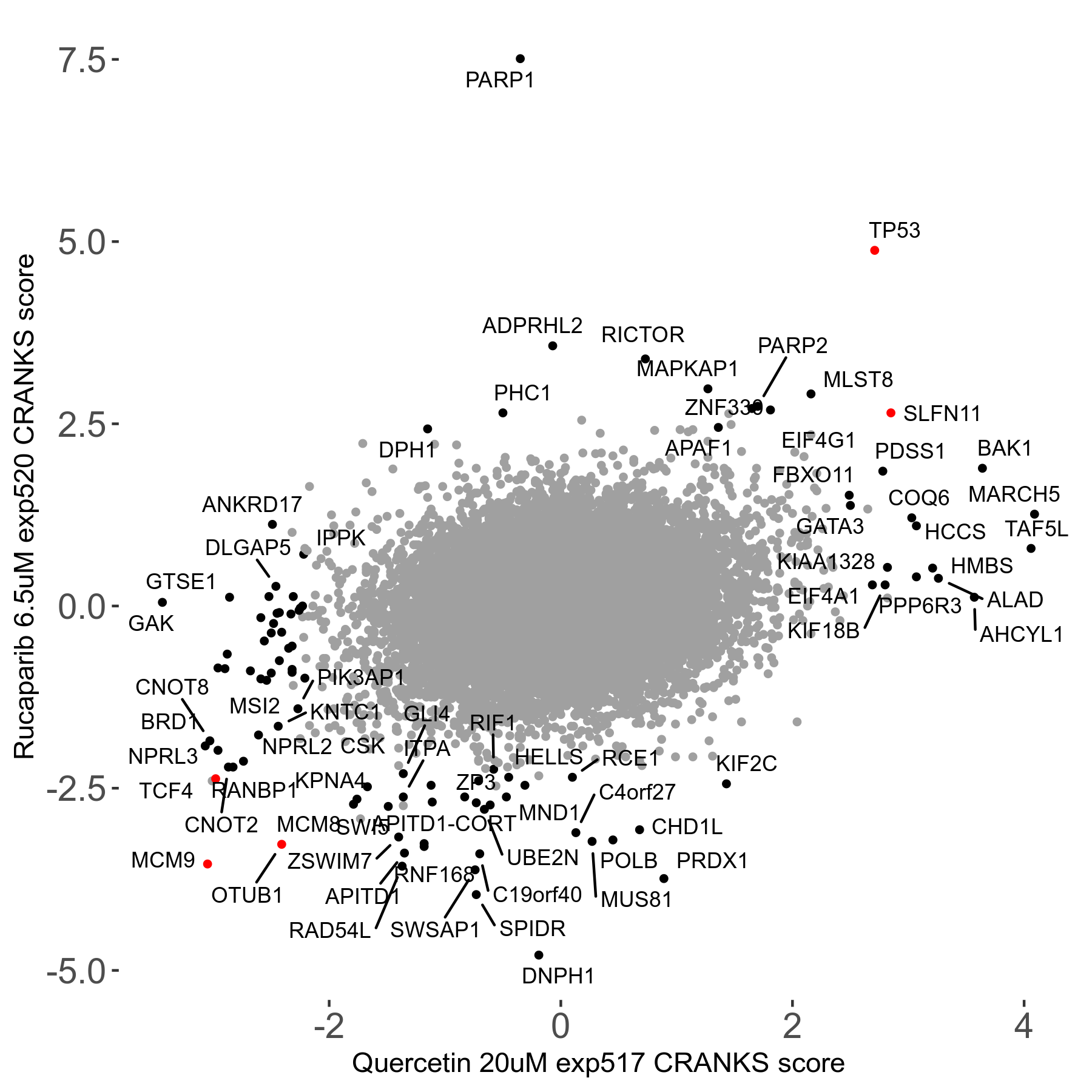
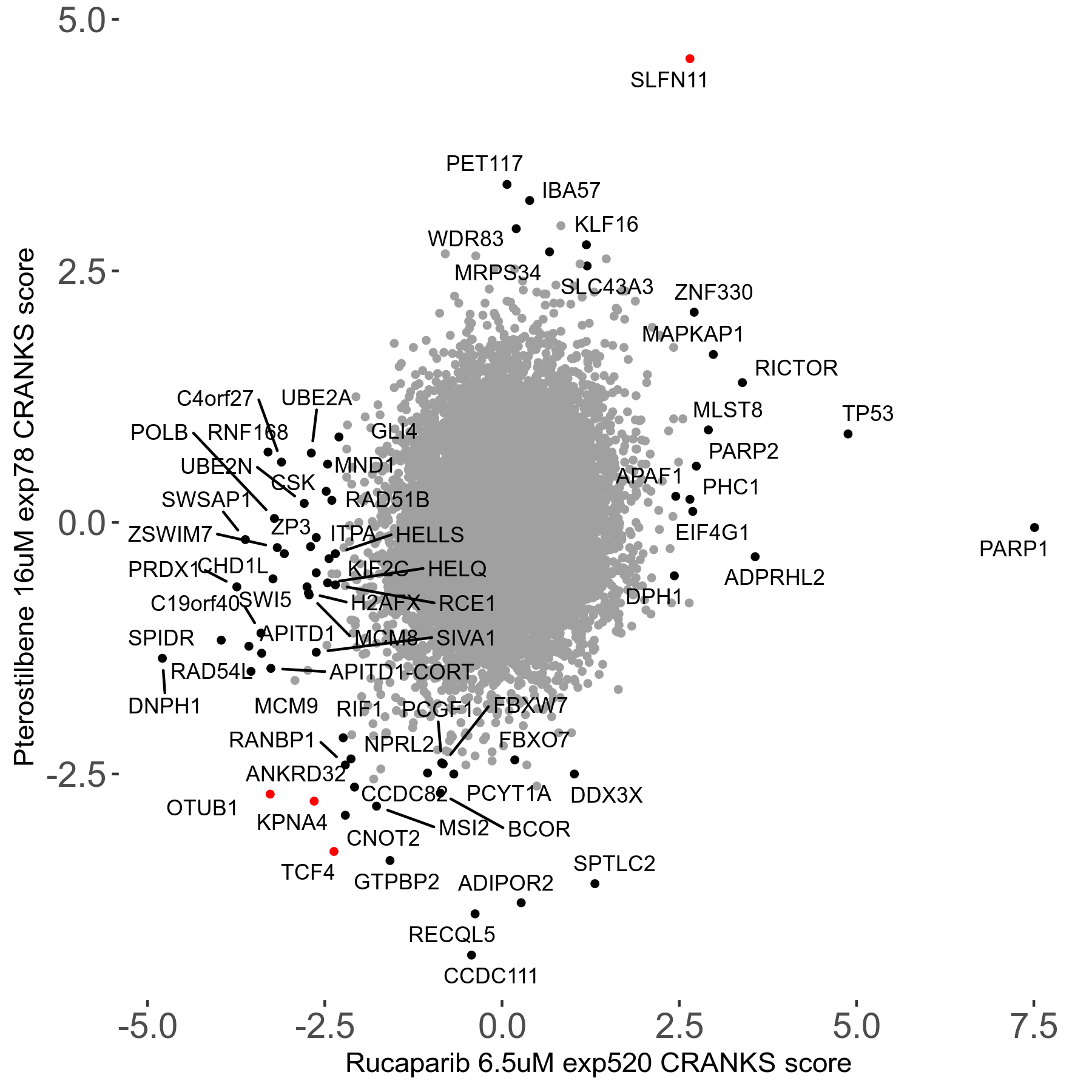
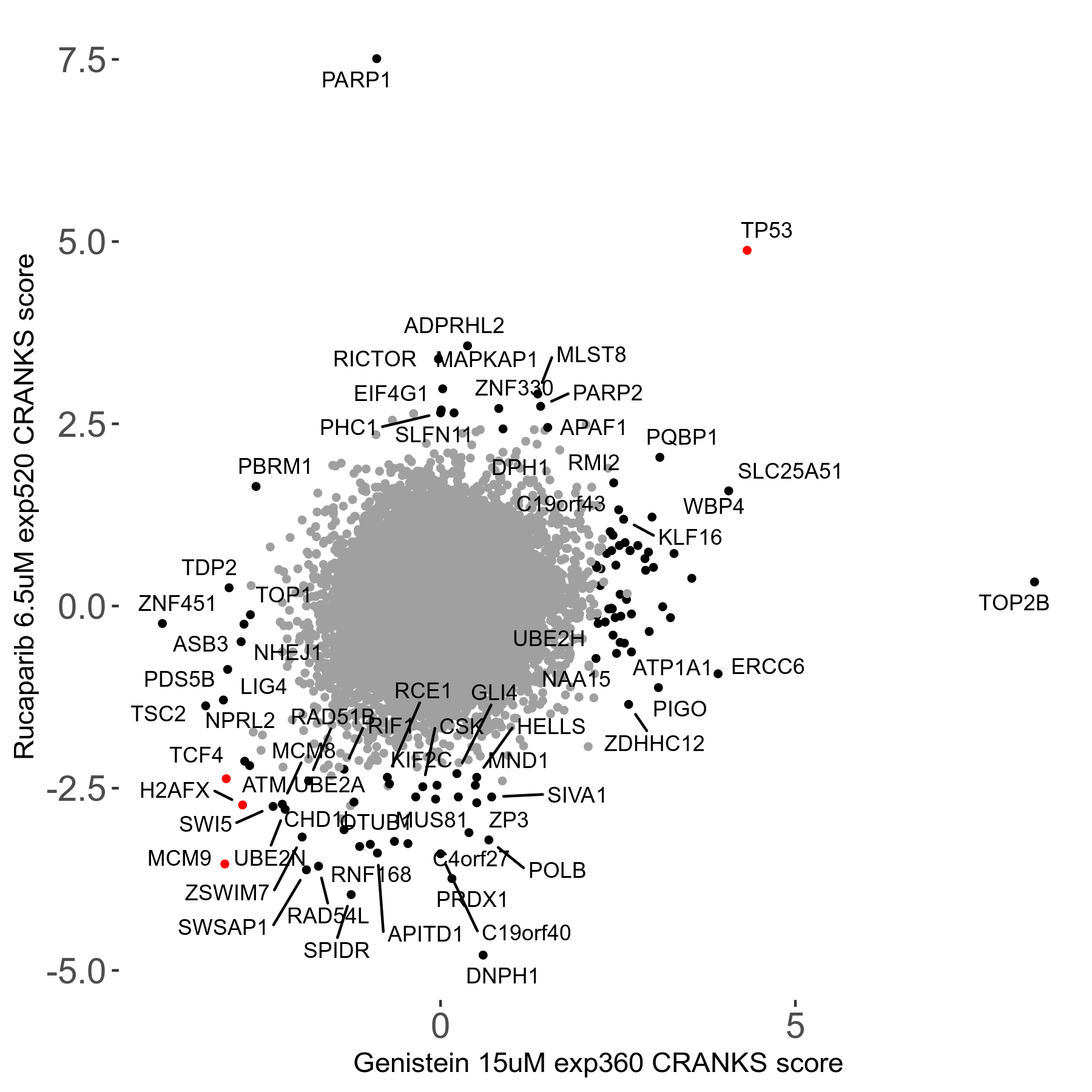
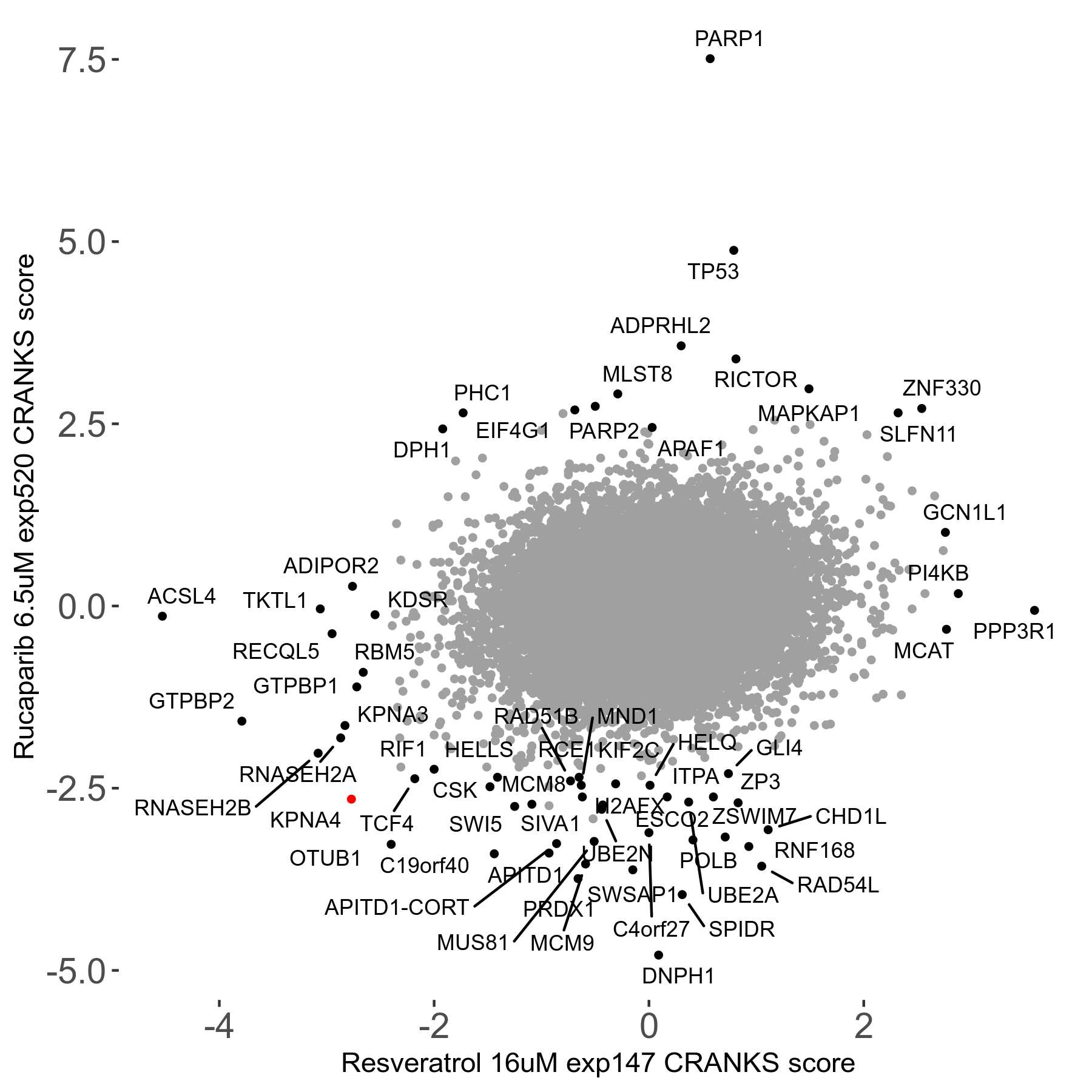
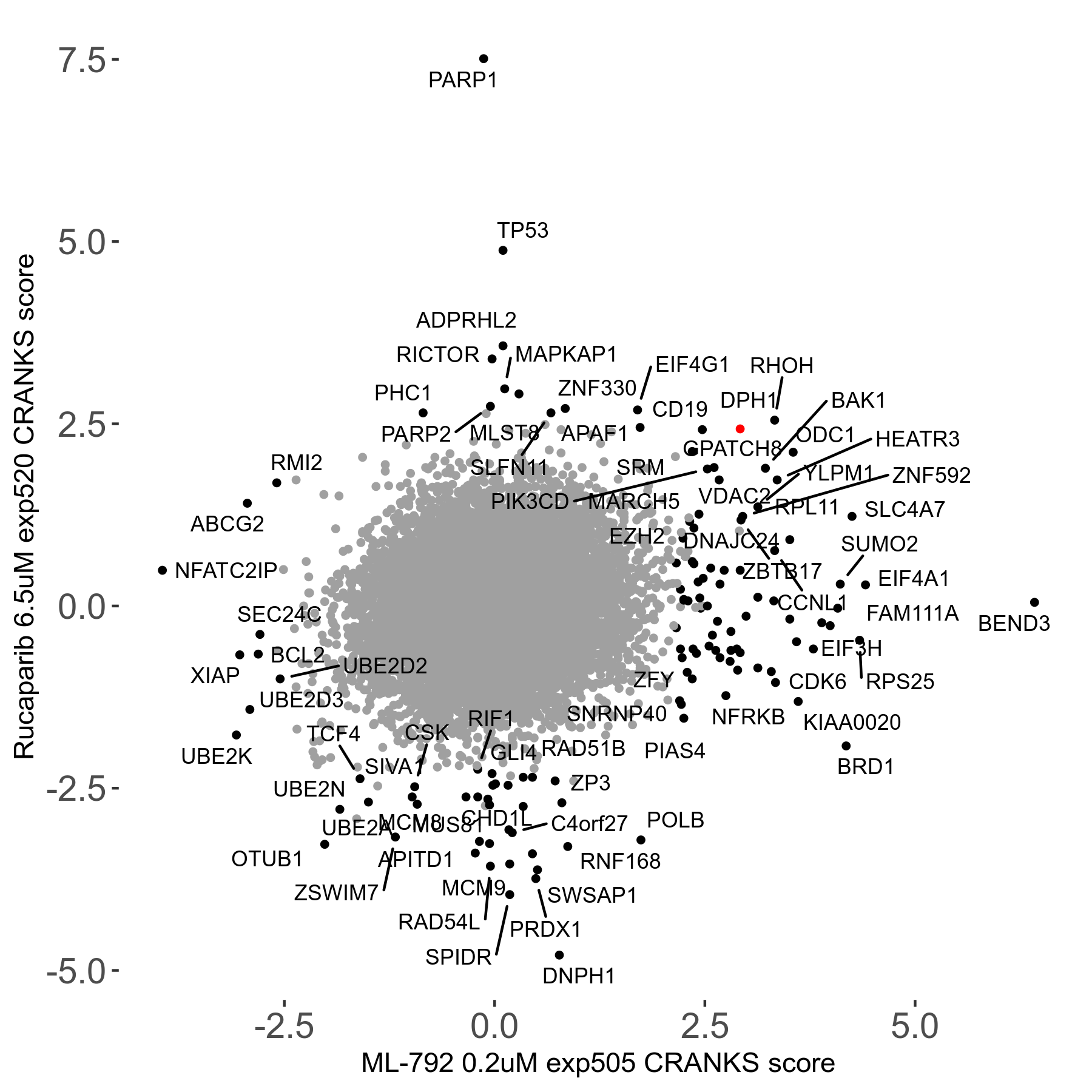
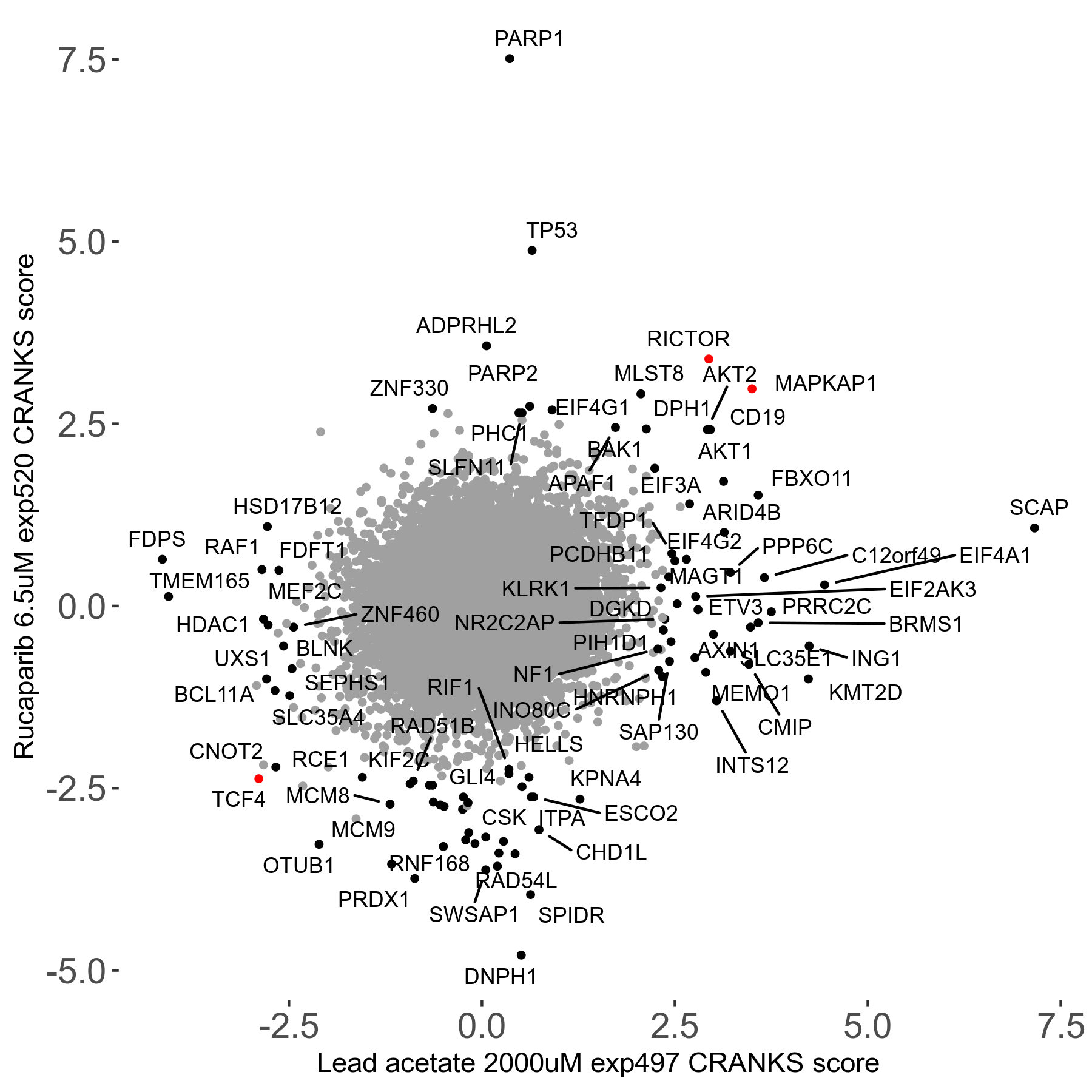
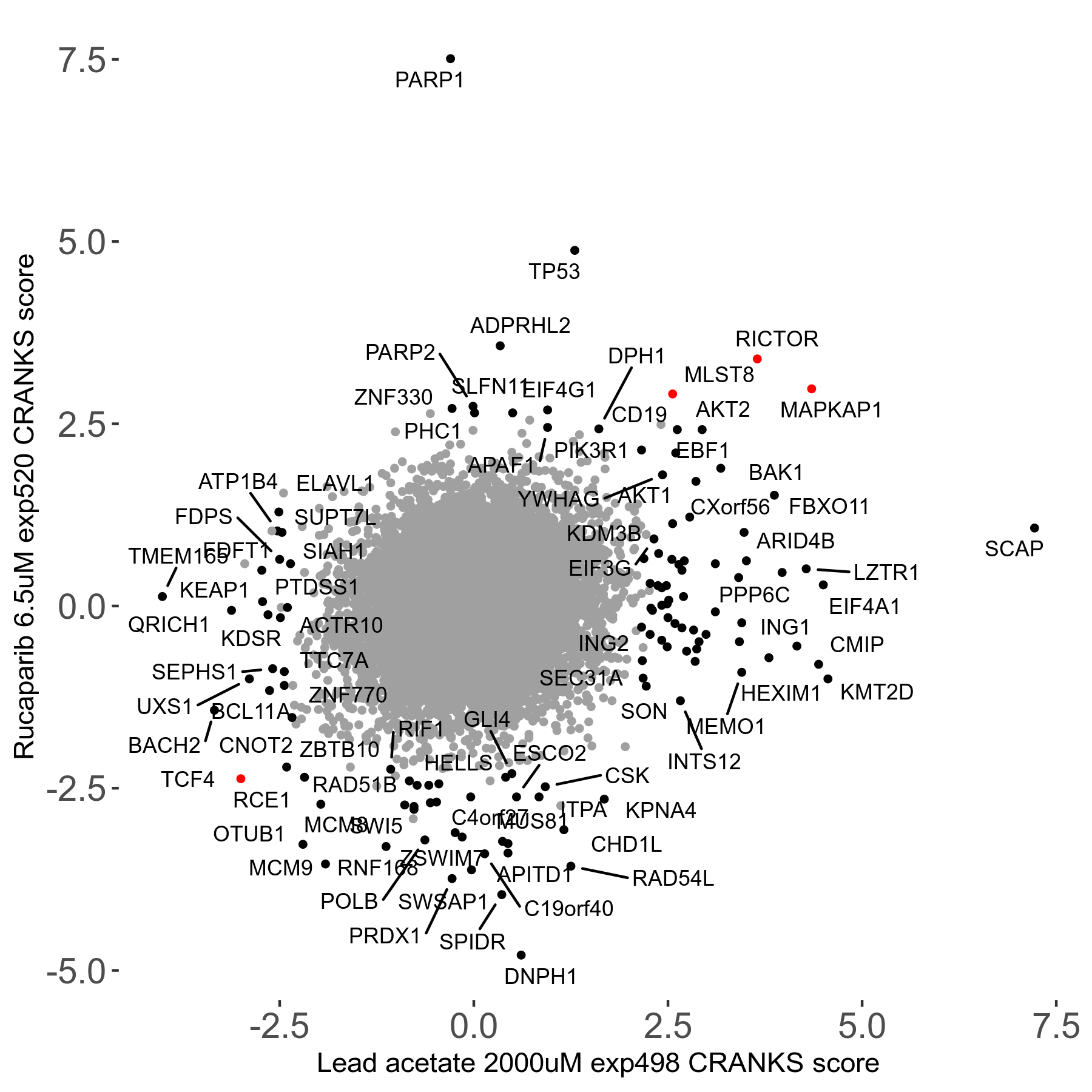
Rucaparib 6.5μM R08 exp520
Mechanism of Action
PARP1/2/3 inhibitor (Clovis), PARP1 mediates single-strand break repair, which can lead to replication-induced double strand breaks to form if not repaired; specifically target cancer with BRCA1, BRCA2 or PALB2 mutations; FDA granted an accelerated approval for use in cases of pretreated advanced ovarian cancer
- Class / Subclass 1: DNA Damage, Repair and Replication / DNA Repair Inhibitor
Technical Notes
Protein References
- PubChem Name: Rucaparib phosphate
- Synonyms: AG-014699 phosphate; PF-01367338 phosphate
- CAS #: 459868-92-9
- PubChem CID: 9931953
- IUPAC: 6-fluoro-2-[4-(methylaminomethyl)phenyl]-3,10-diazatricyclo[6.4.1.04,13]trideca-1,4,6,8(13)-tetraen-9-one;phosphoric acid
- INCHI Name: InChI=1S/C19H18FN3O.H3O4P/c1-21-10-11-2-4-12(5-3-11)18-14-6-7-22-19(24)15-8-13(20)9-16(23-18)17(14)15;1-5(2,3)4/h2-5,8-9,21,23H,6-7,10H2,1H3,(H,22,24);(H3,1,2,3,4)
- INCHI Key: FCCGJTKEKXUBFZ-UHFFFAOYSA-N
- Molecular Weight: 421.4
- Canonical SMILES: CNCC1=CC=C(C=C1)C2=C3CCNC(=O)C4=C3C(=CC(=C4)F)N2.OP(=O)(O)O
- Isomeric SMILES: N/A
- Molecular Formula: C19H21FN3O5P
Protein Supplier
- Supplier Name: Med Chem Express
- Catalog #: HY-10617
- Lot #: 29562
Protein Characterization
- HRMS (ESI-TOF) m/z: (M+H)+ Calcd for C19H18FN3O 324.15067; found 324.15184
Dose Response Curve
- Platform ID: Rucaparib
- Min: 4.0175; Max: 99.5587

| IC | Concentration (µM) |
|---|---|
| IC10 | 3.6040 |
| IC20 | 5.1380 |
| IC30 | 6.5040 |
| IC40 | 7.8900 |
| IC50 | 9.4220 |
| IC60 | 11.2500 |
| IC70 | 13.6500 |
| IC80 | 17.2800 |
| IC90 | 24.6300 |
Screen Summary
- Round: 08
- Dose: 6.5µM
- Days of incubation: 8
- Doublings: 3.6
- Numbers of reads: 16082760
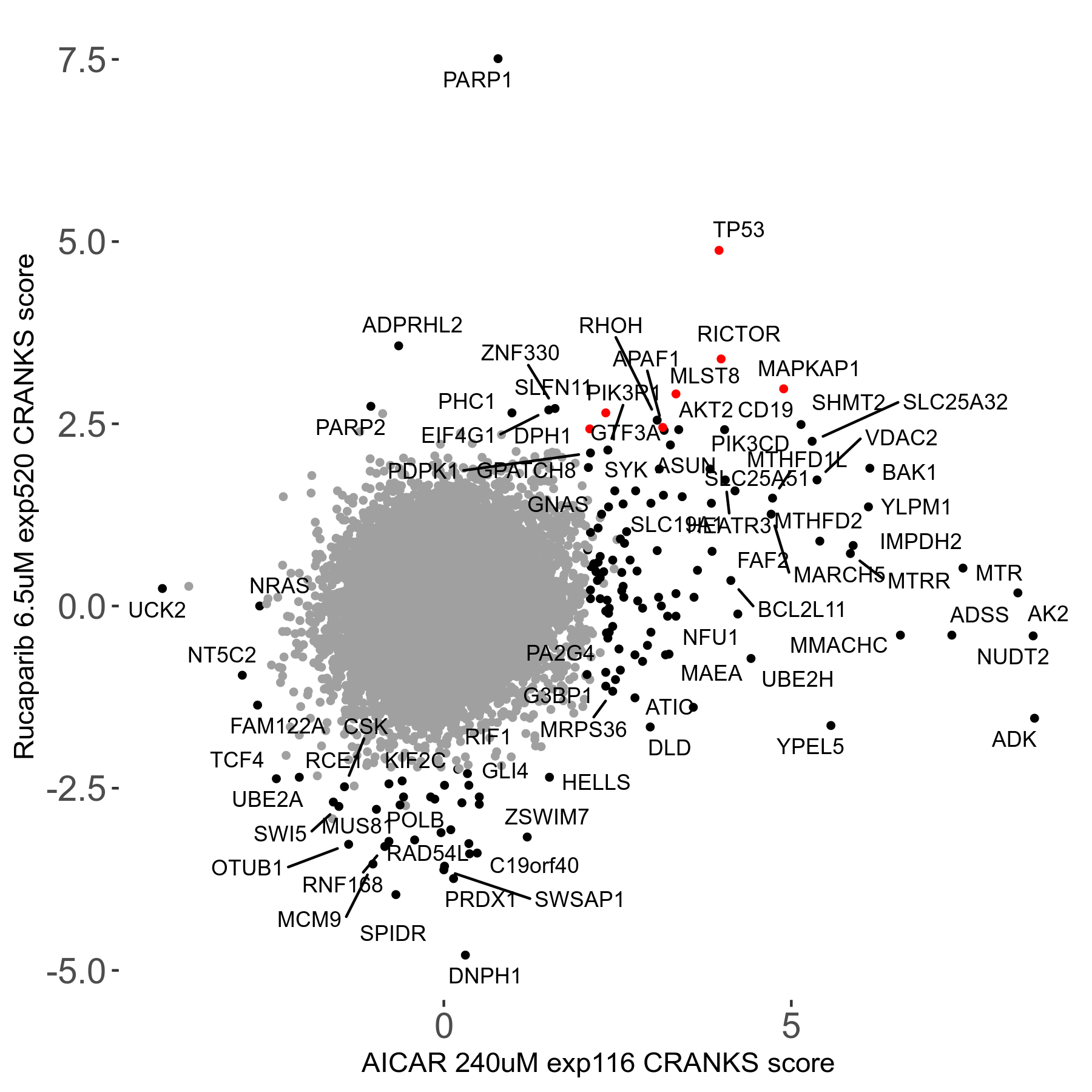
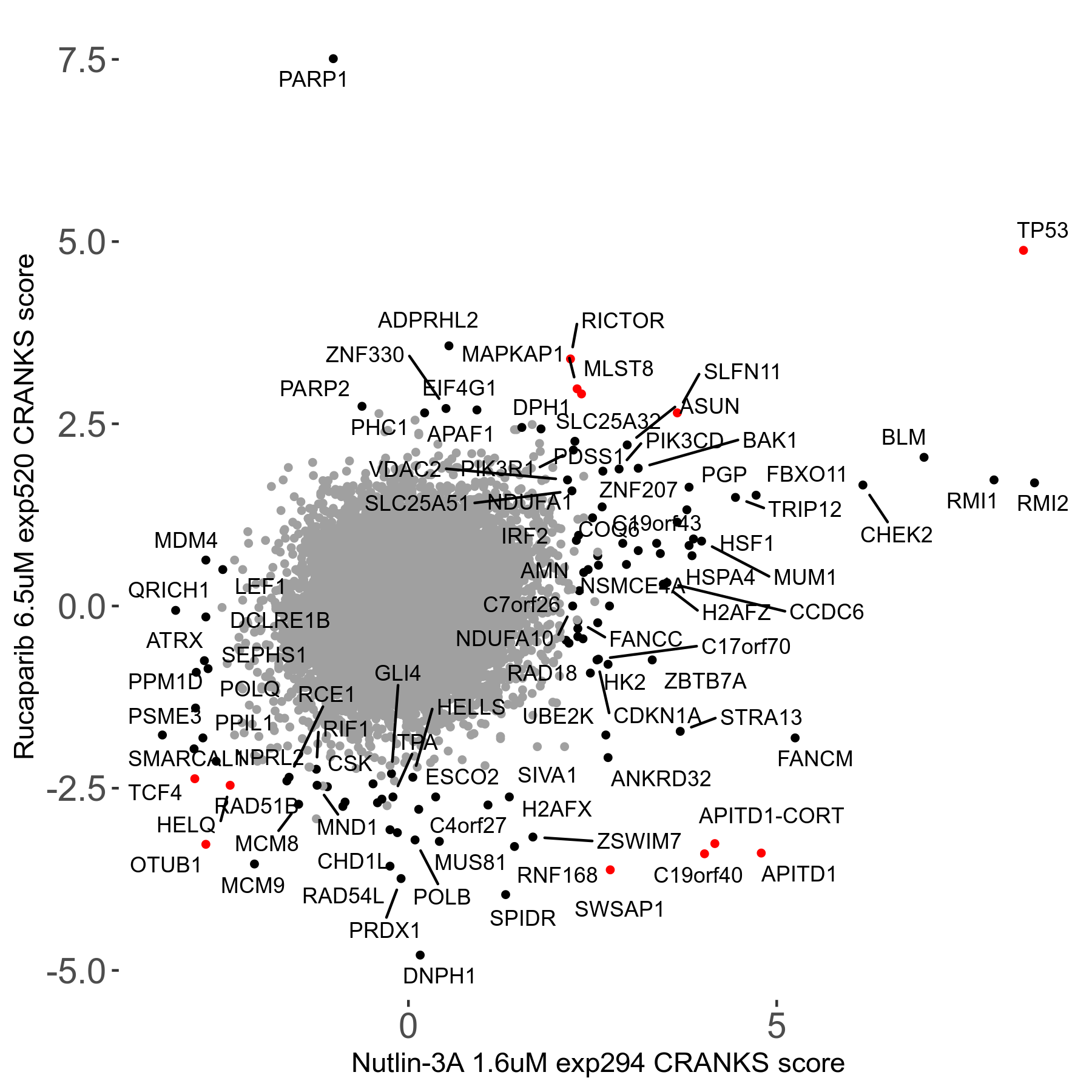
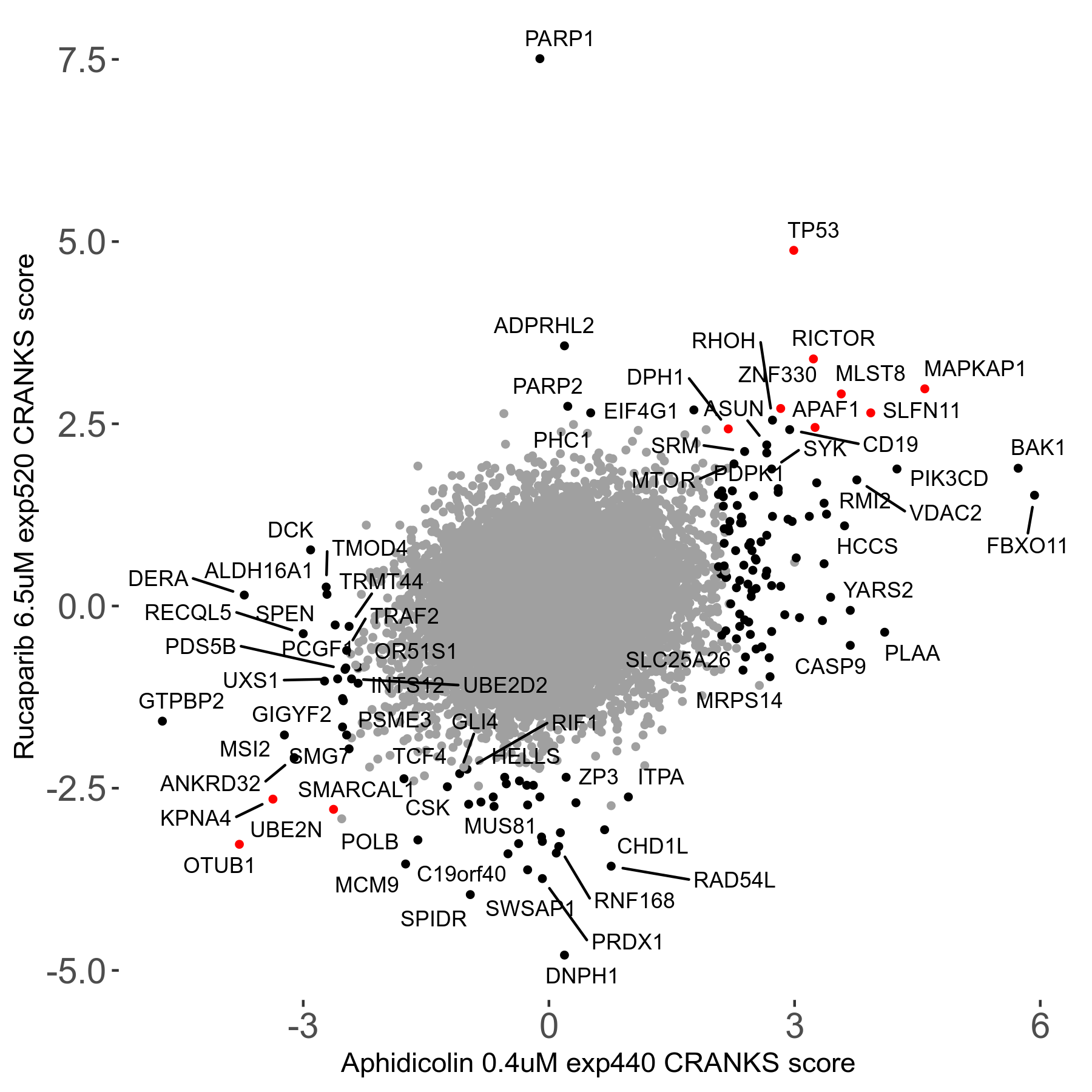
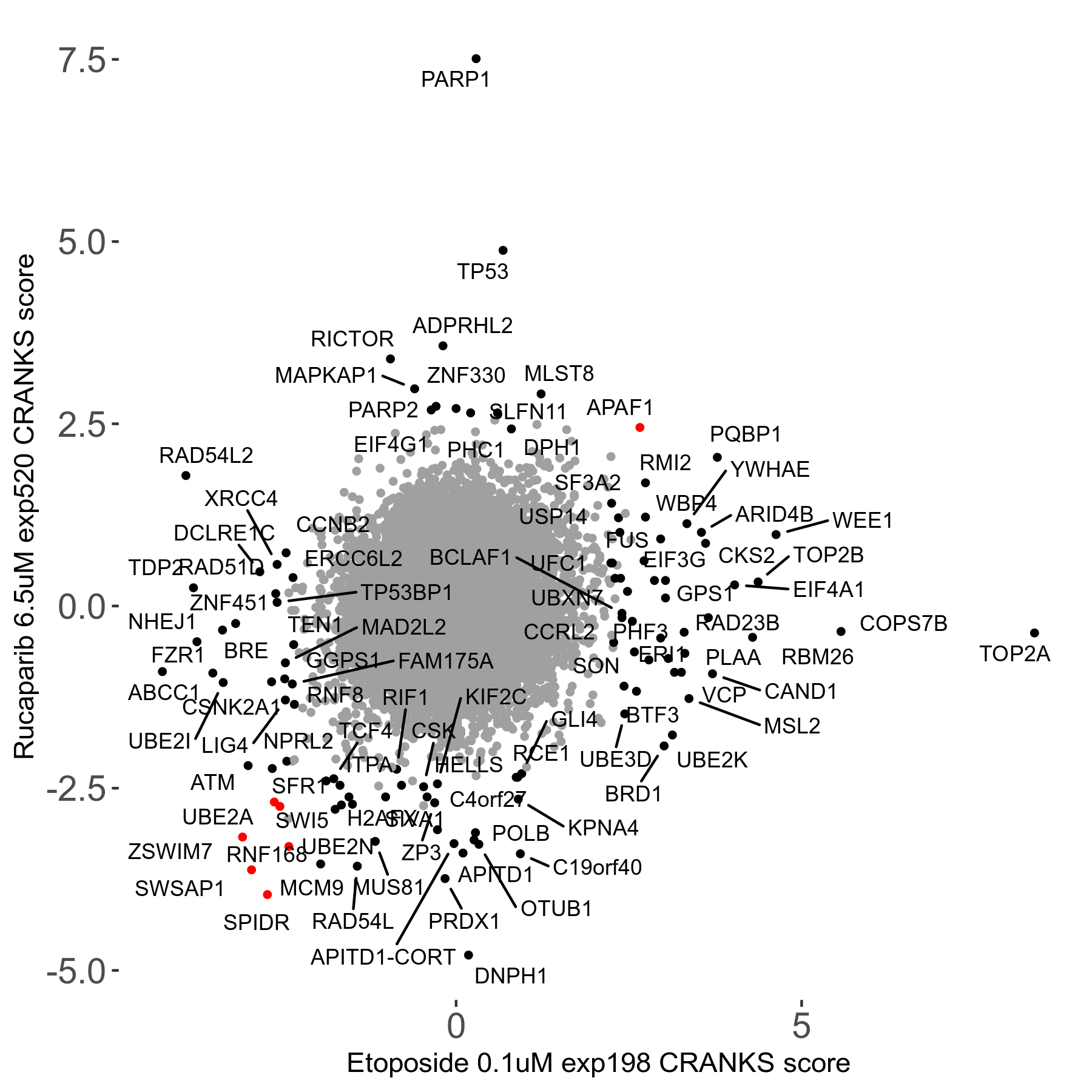
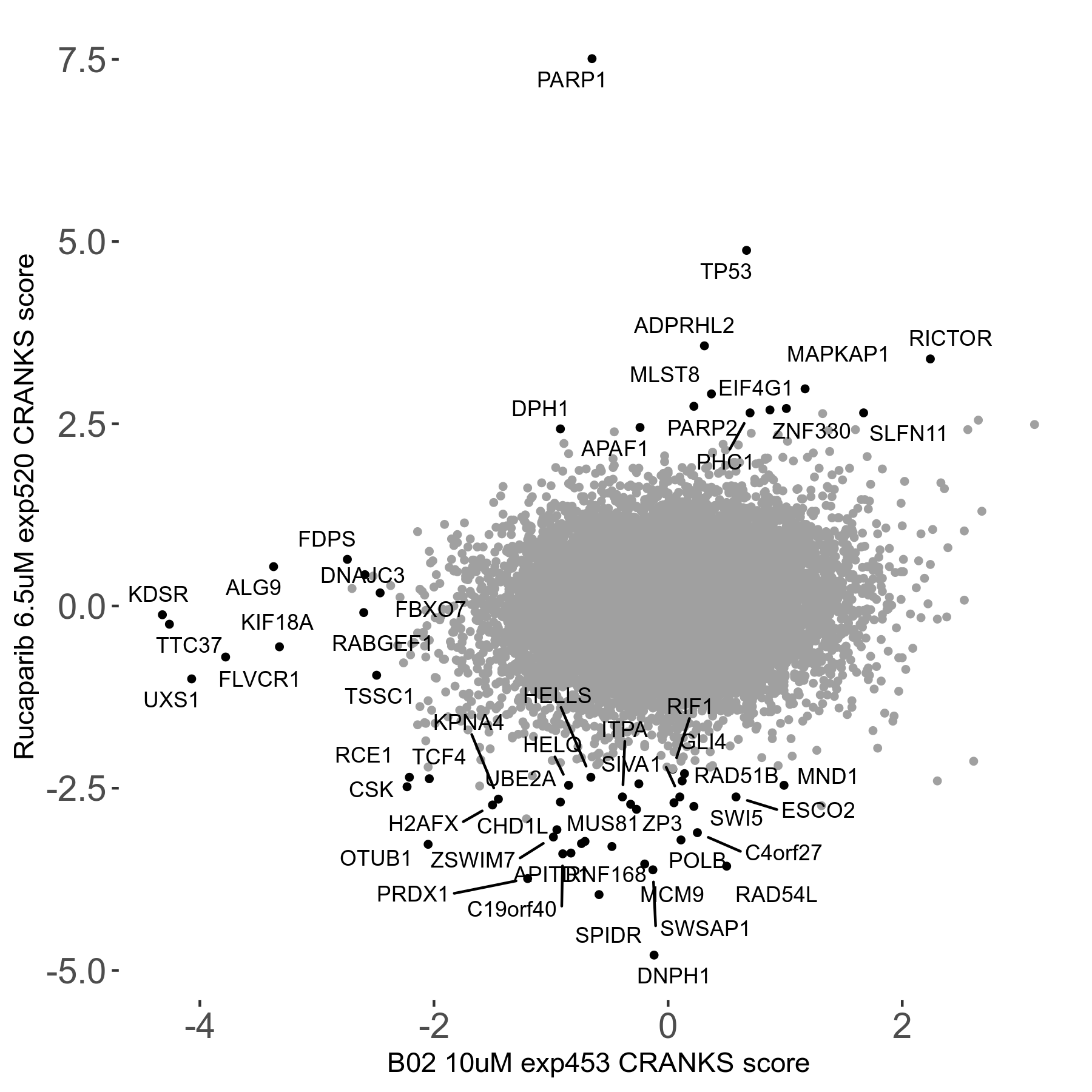
Screen Results
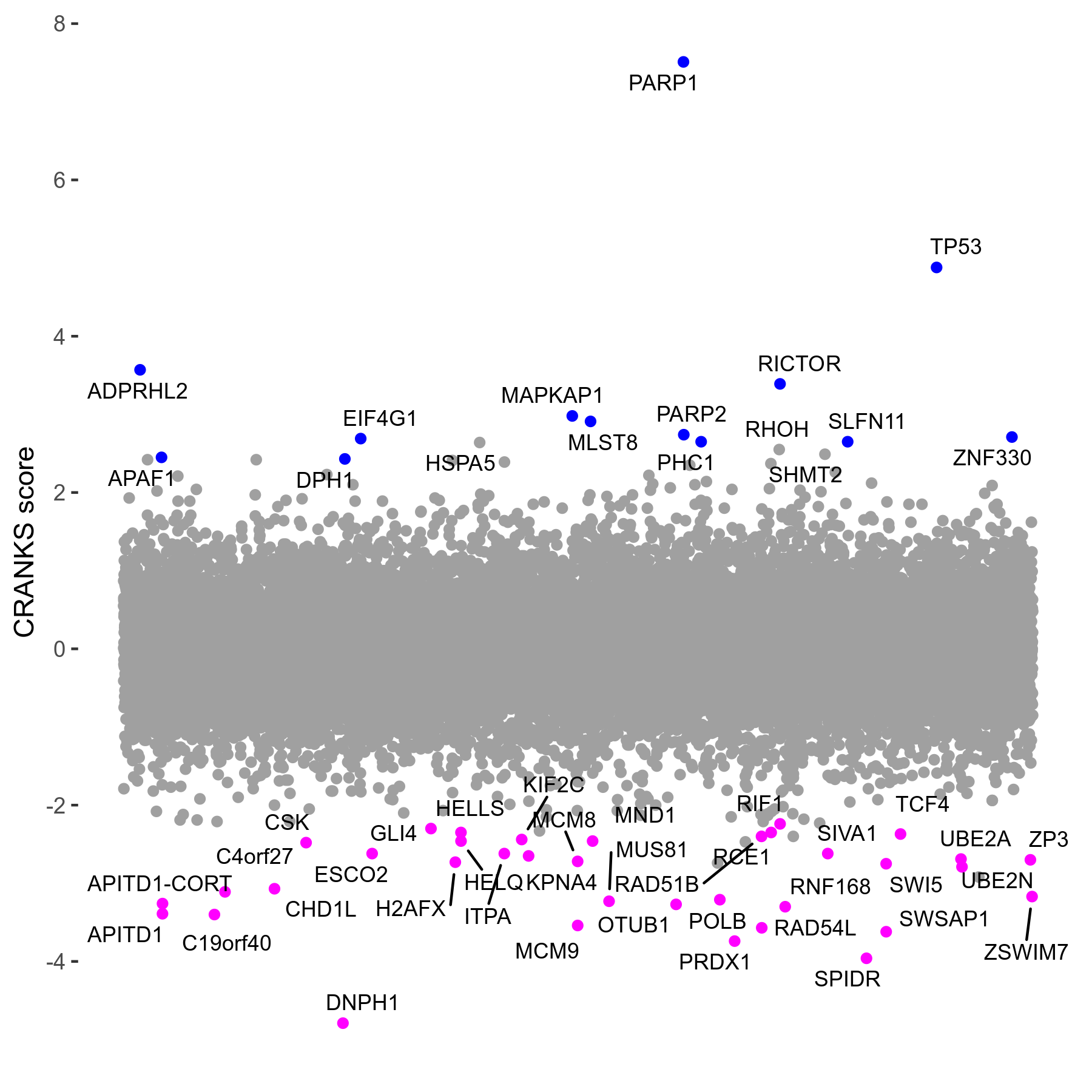
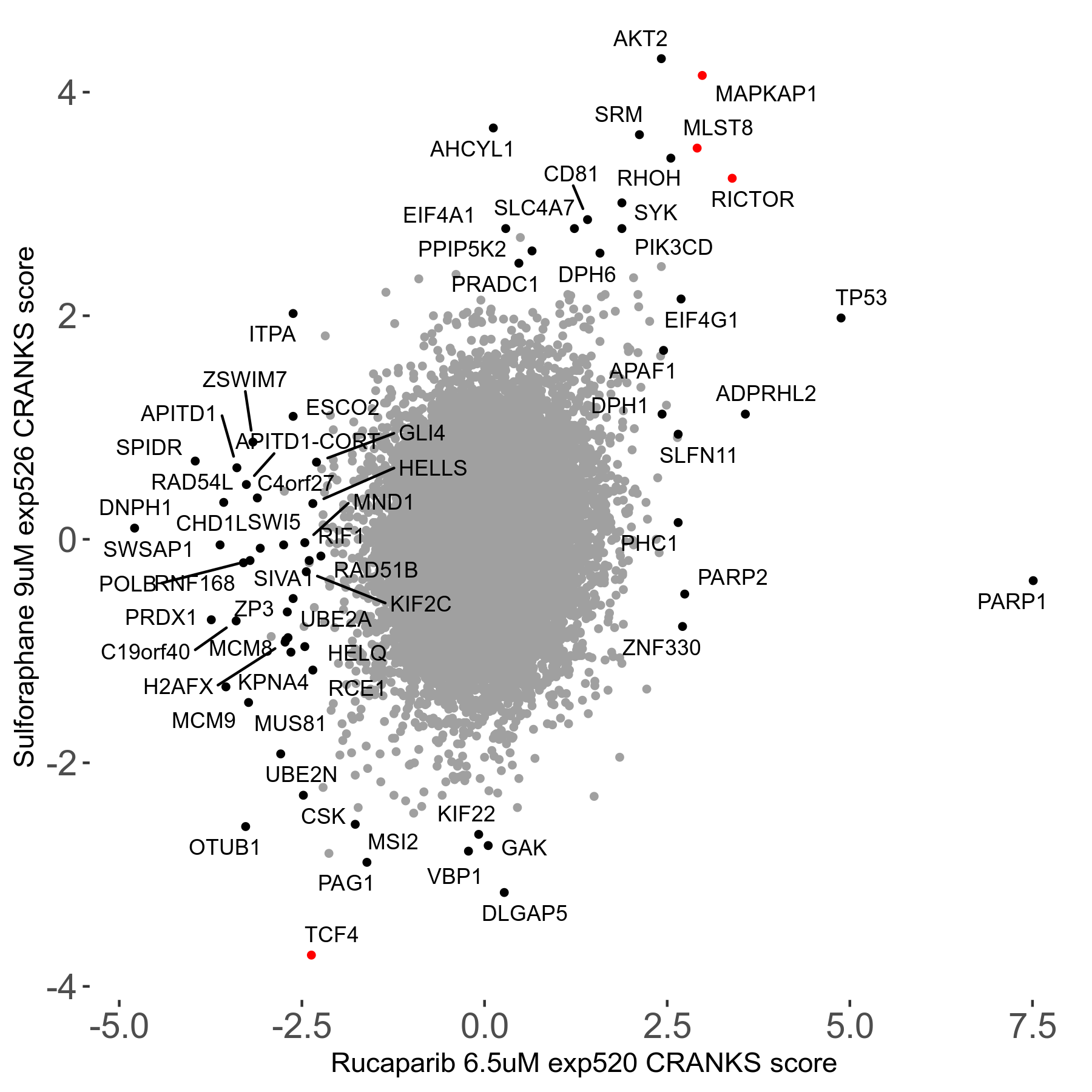
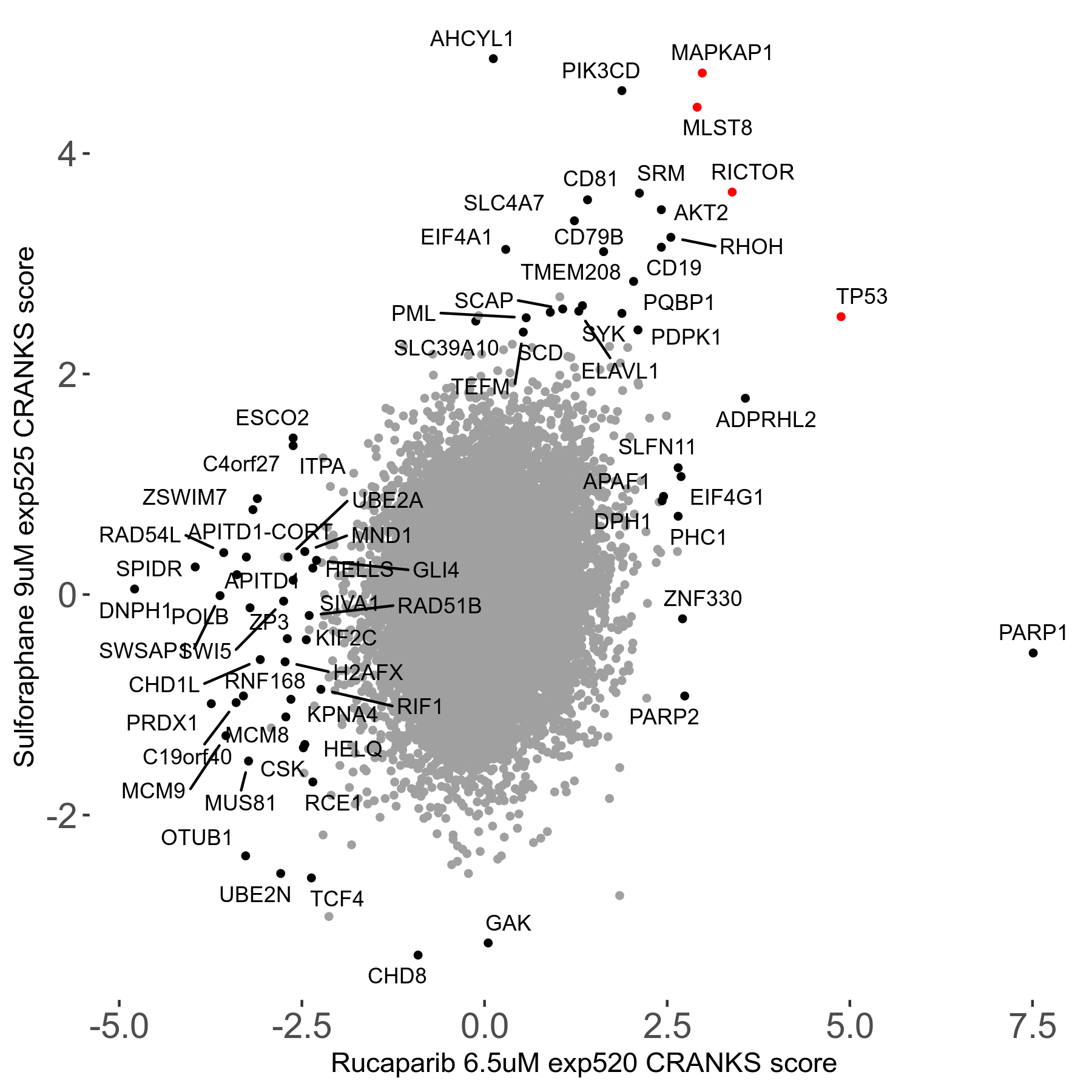
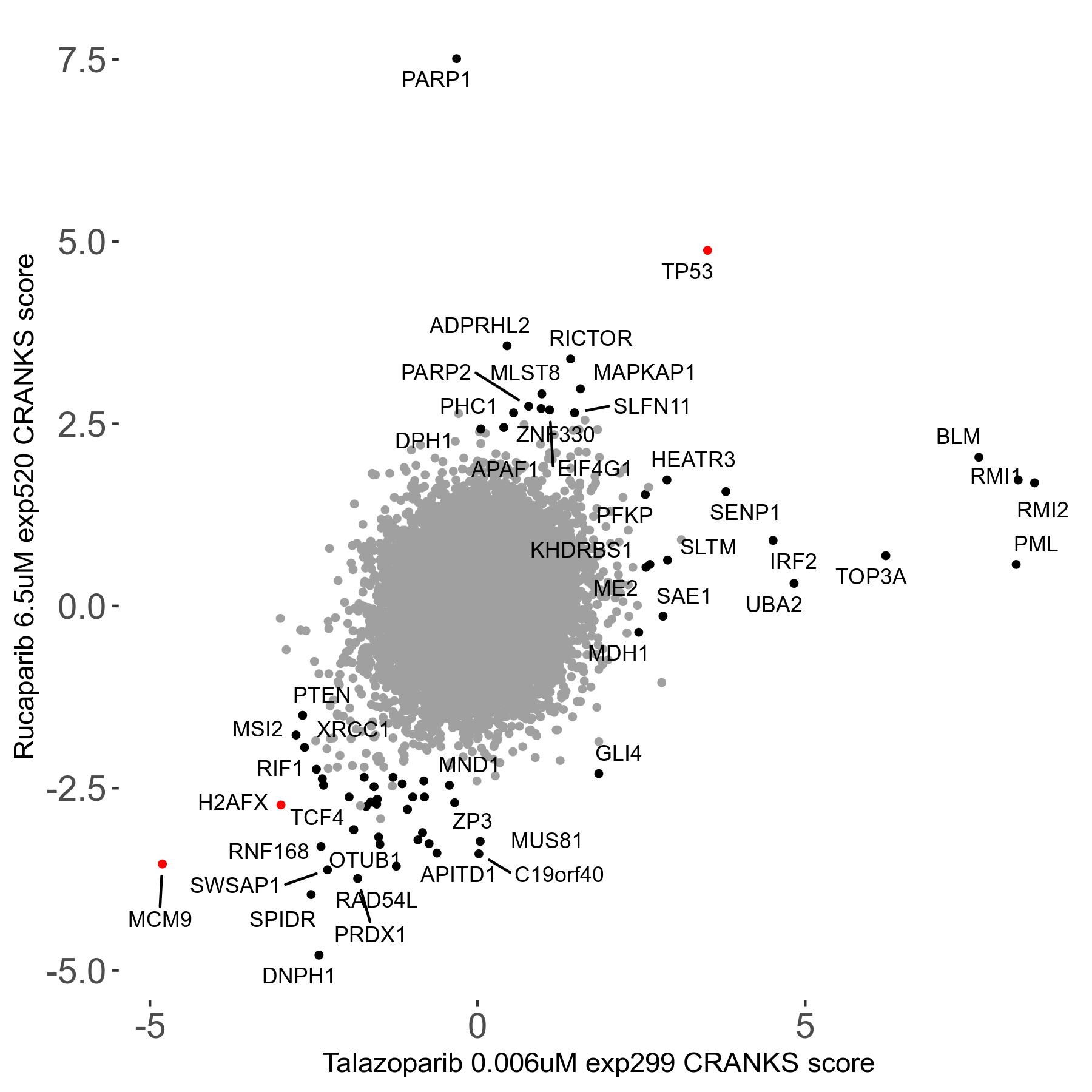
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 36/13 | Scores |