Table of Contents
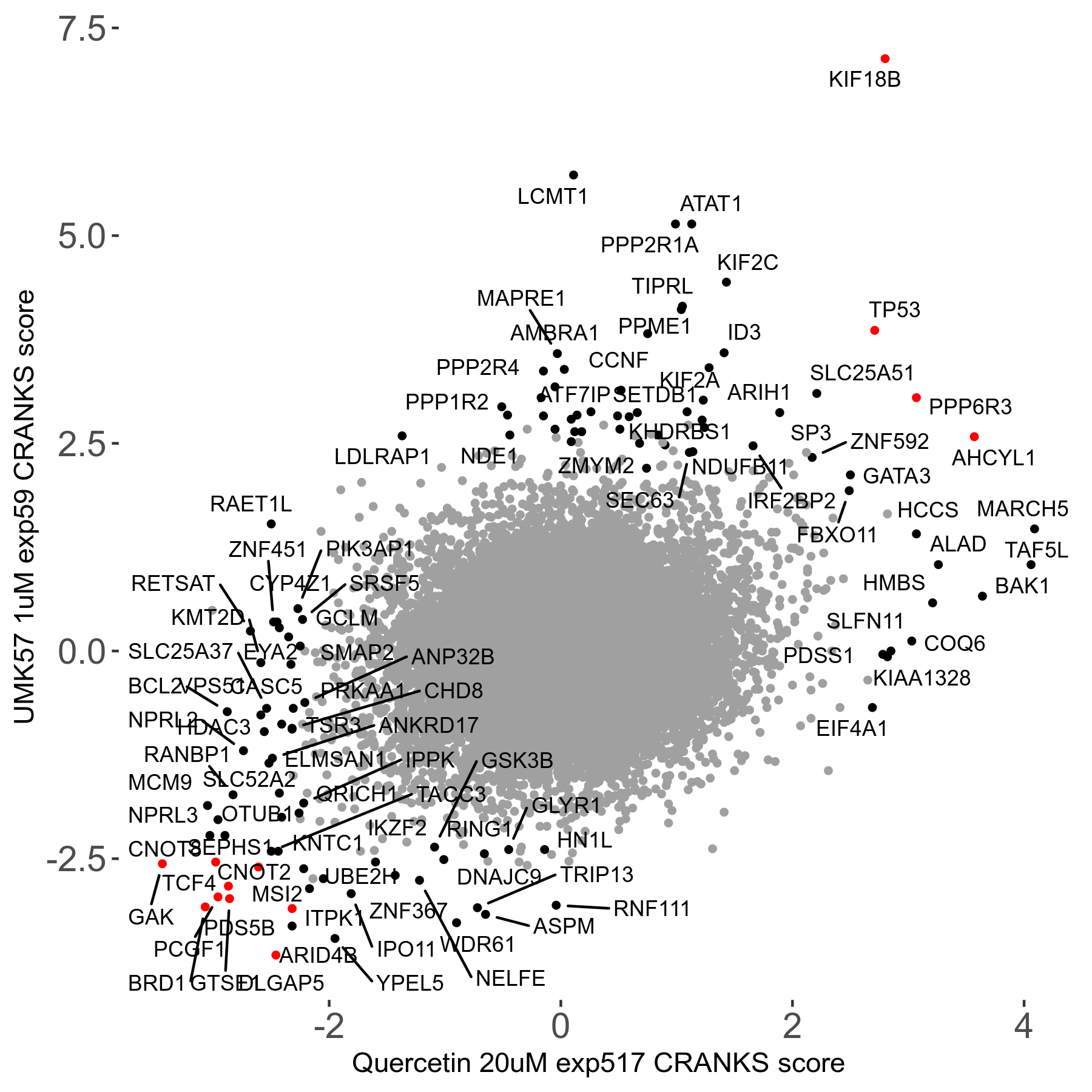
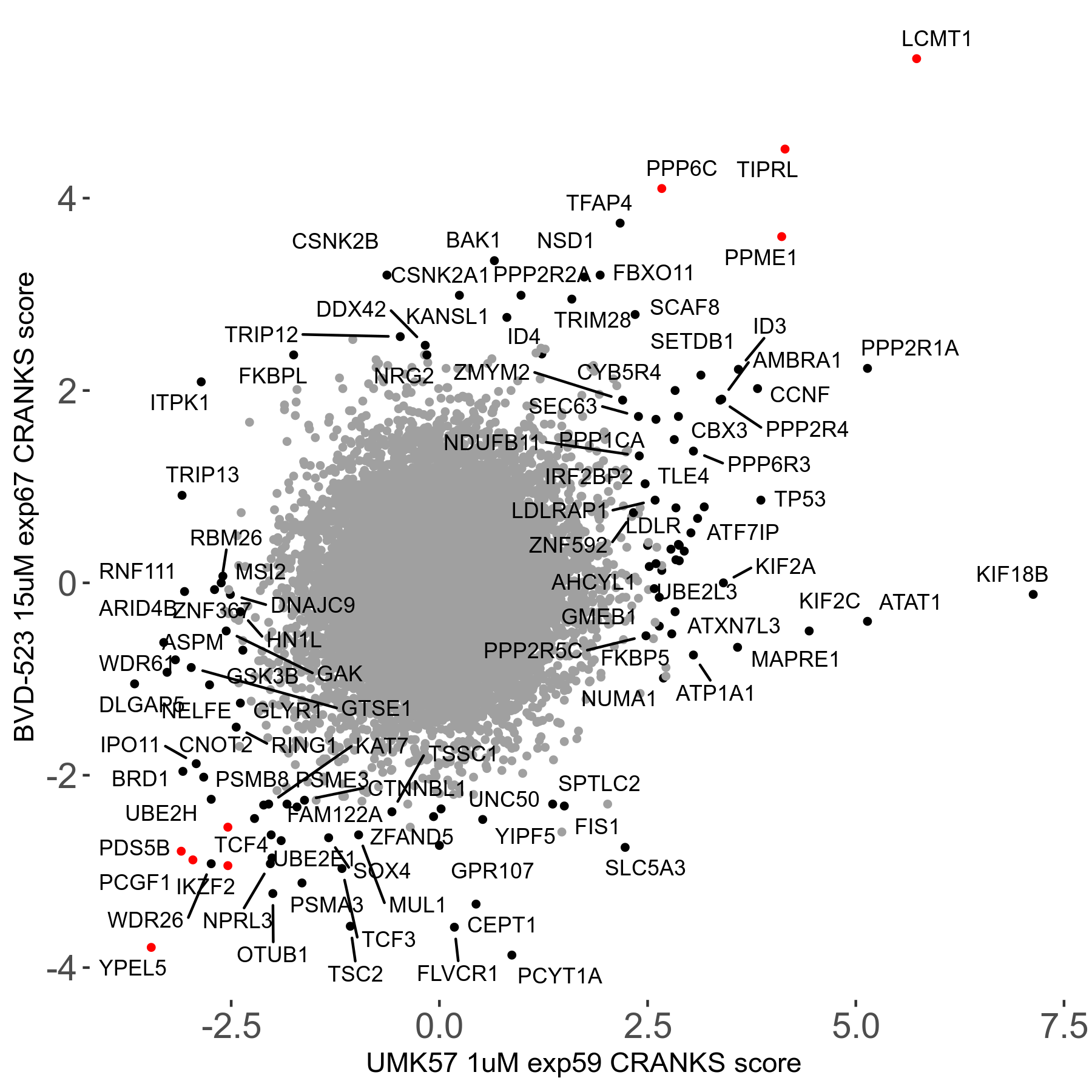
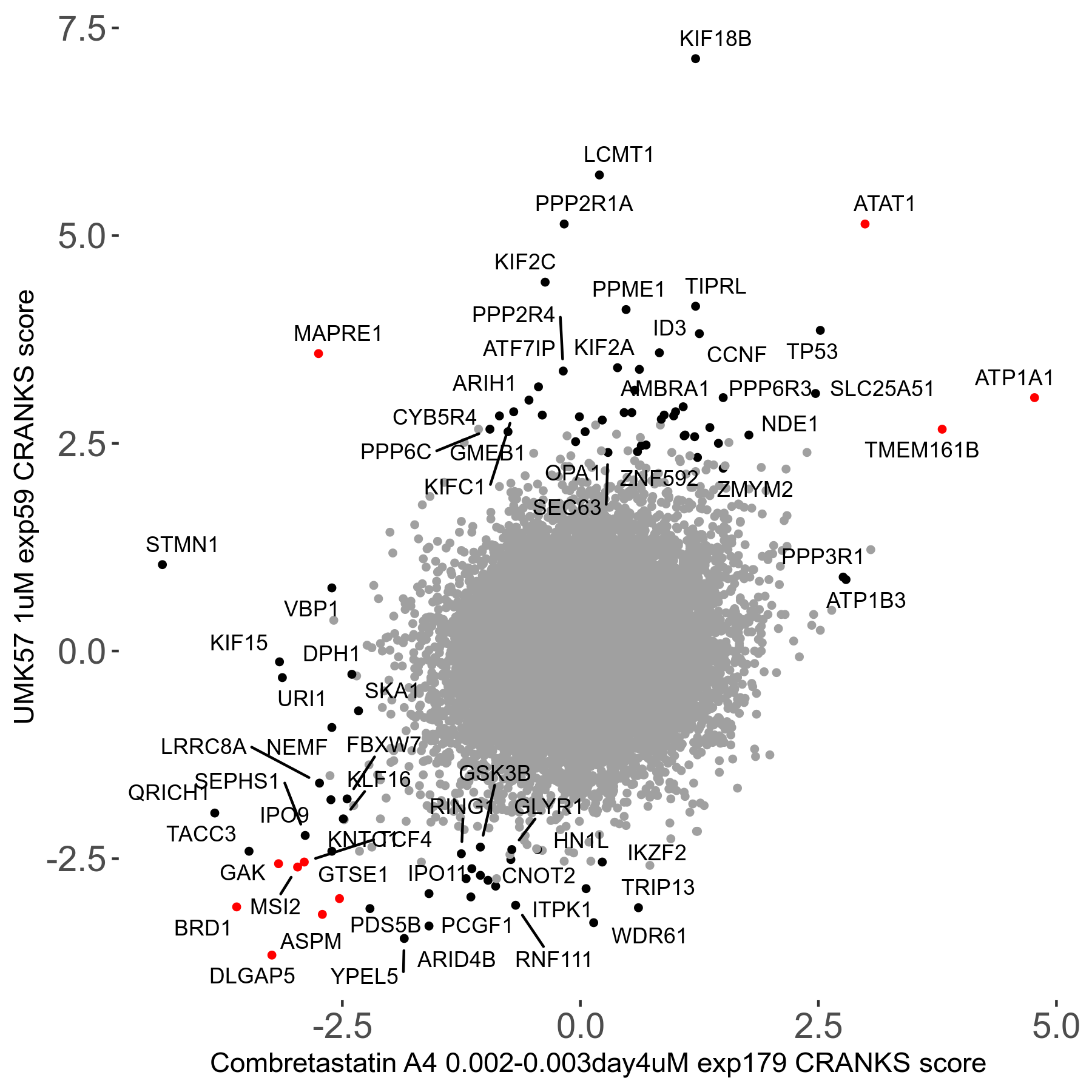
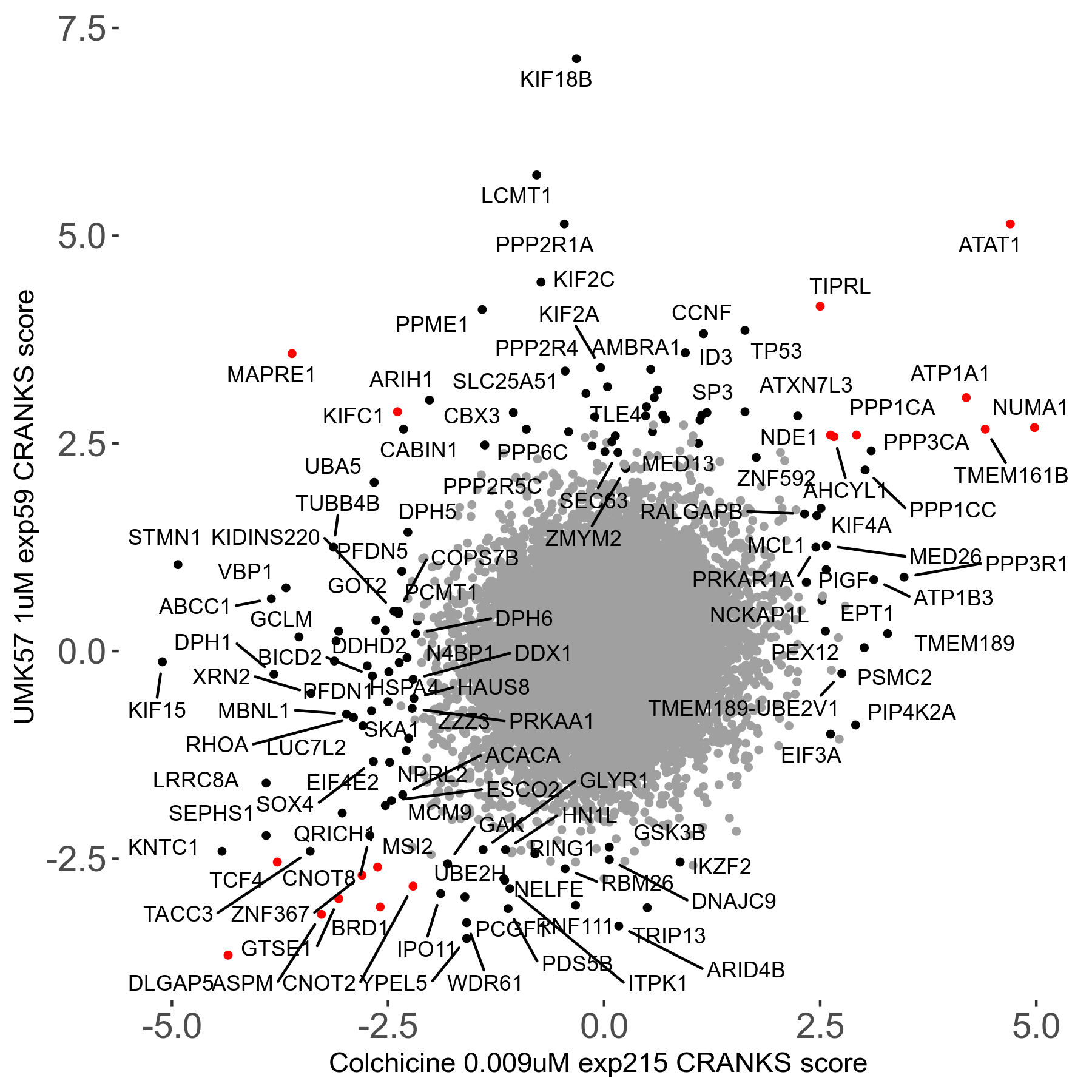
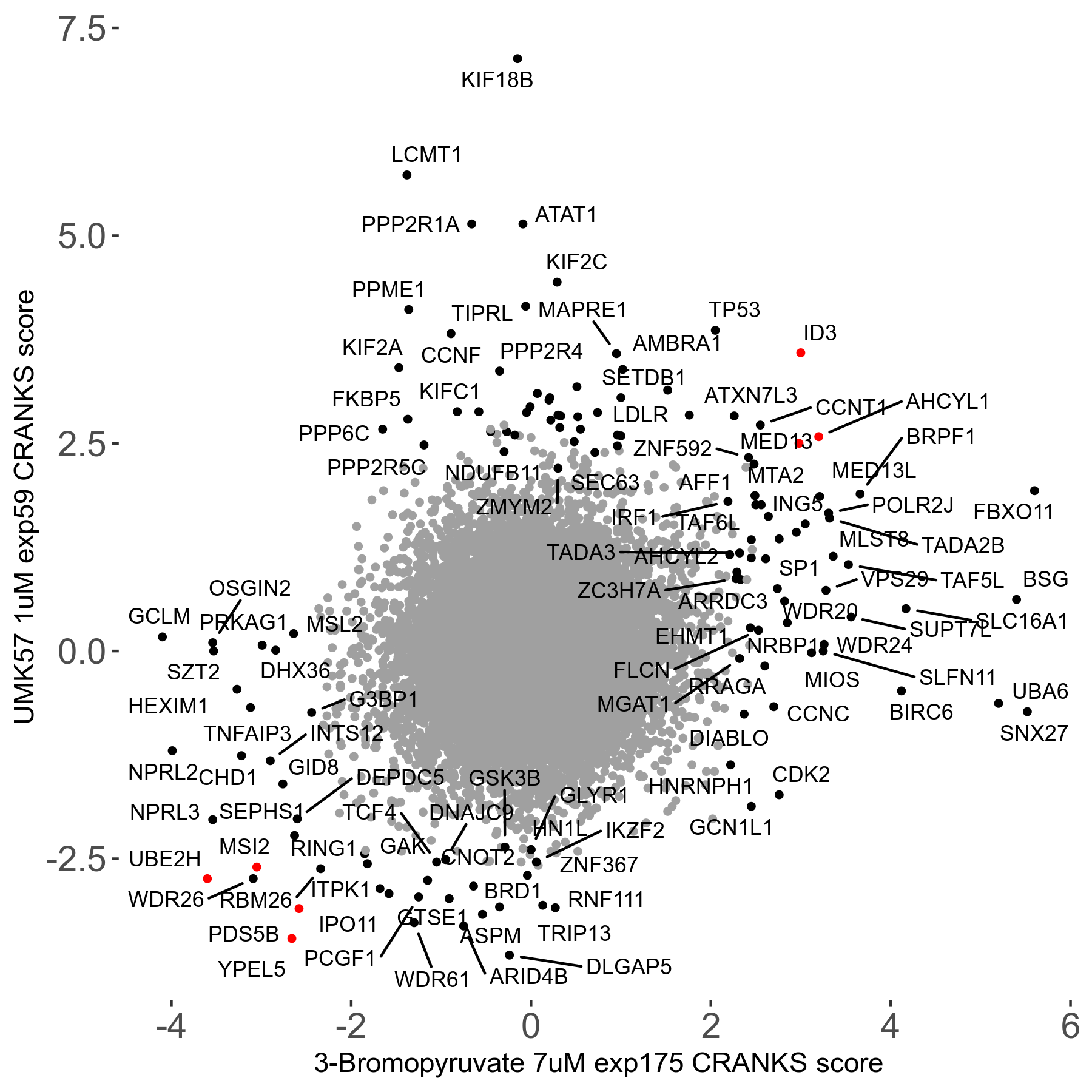
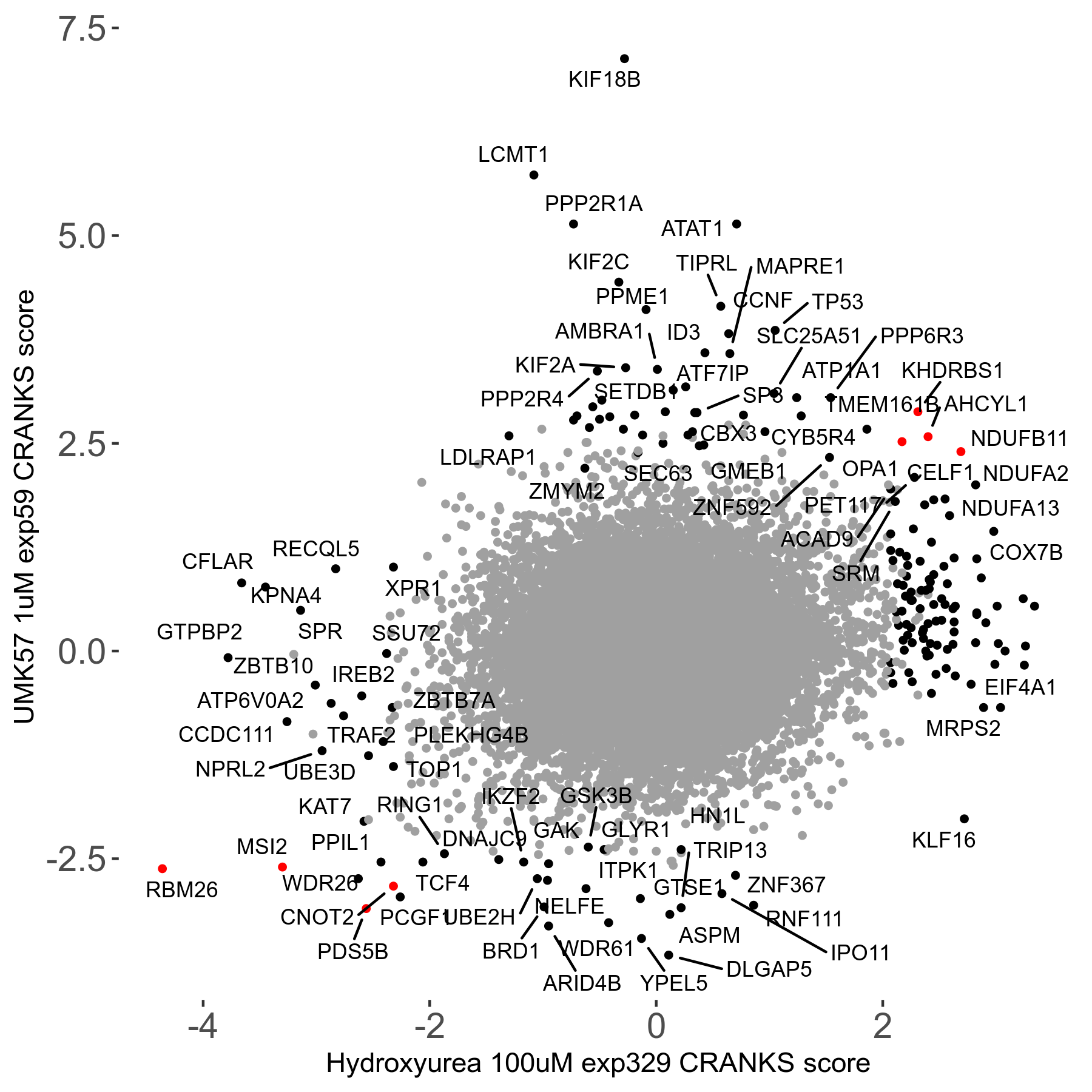
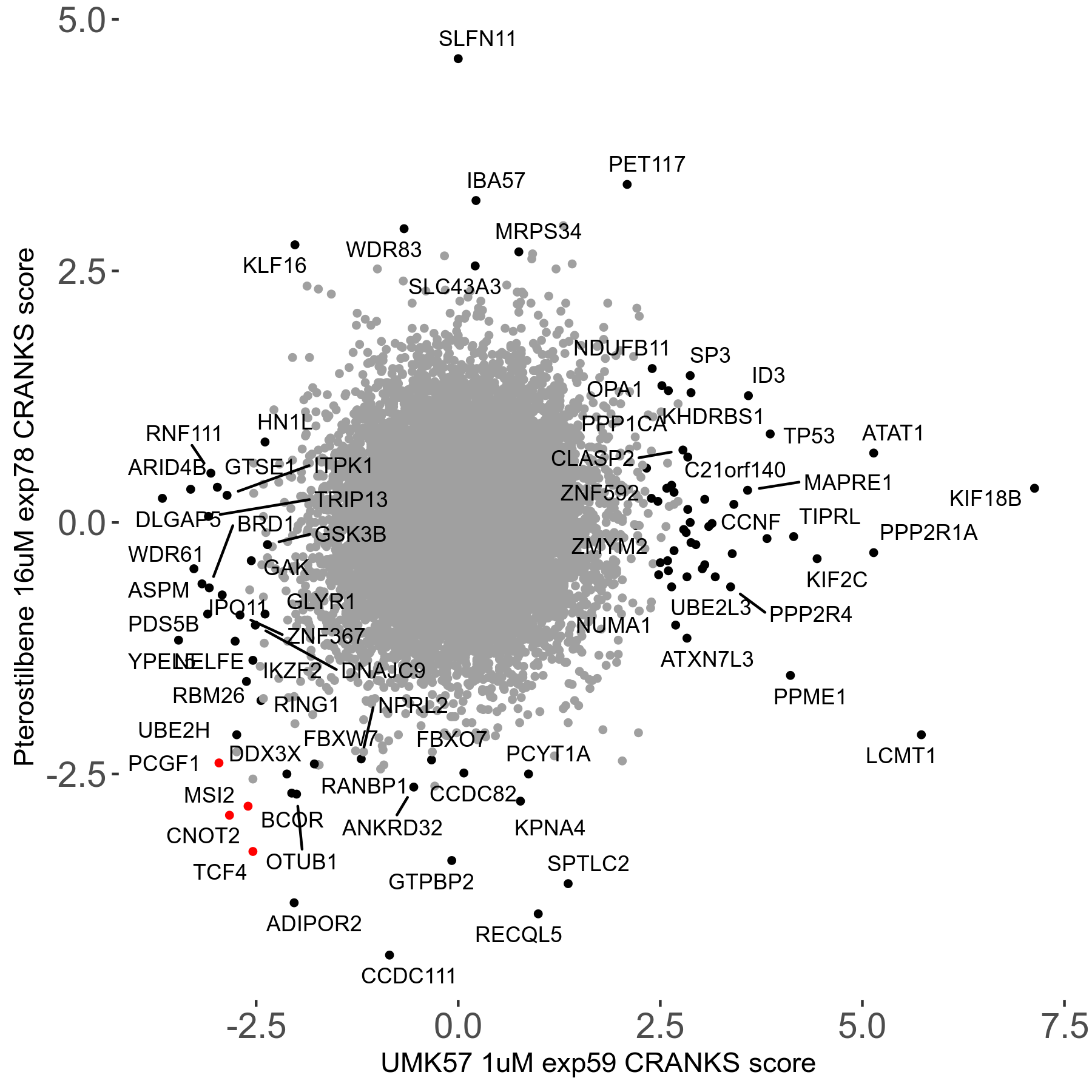
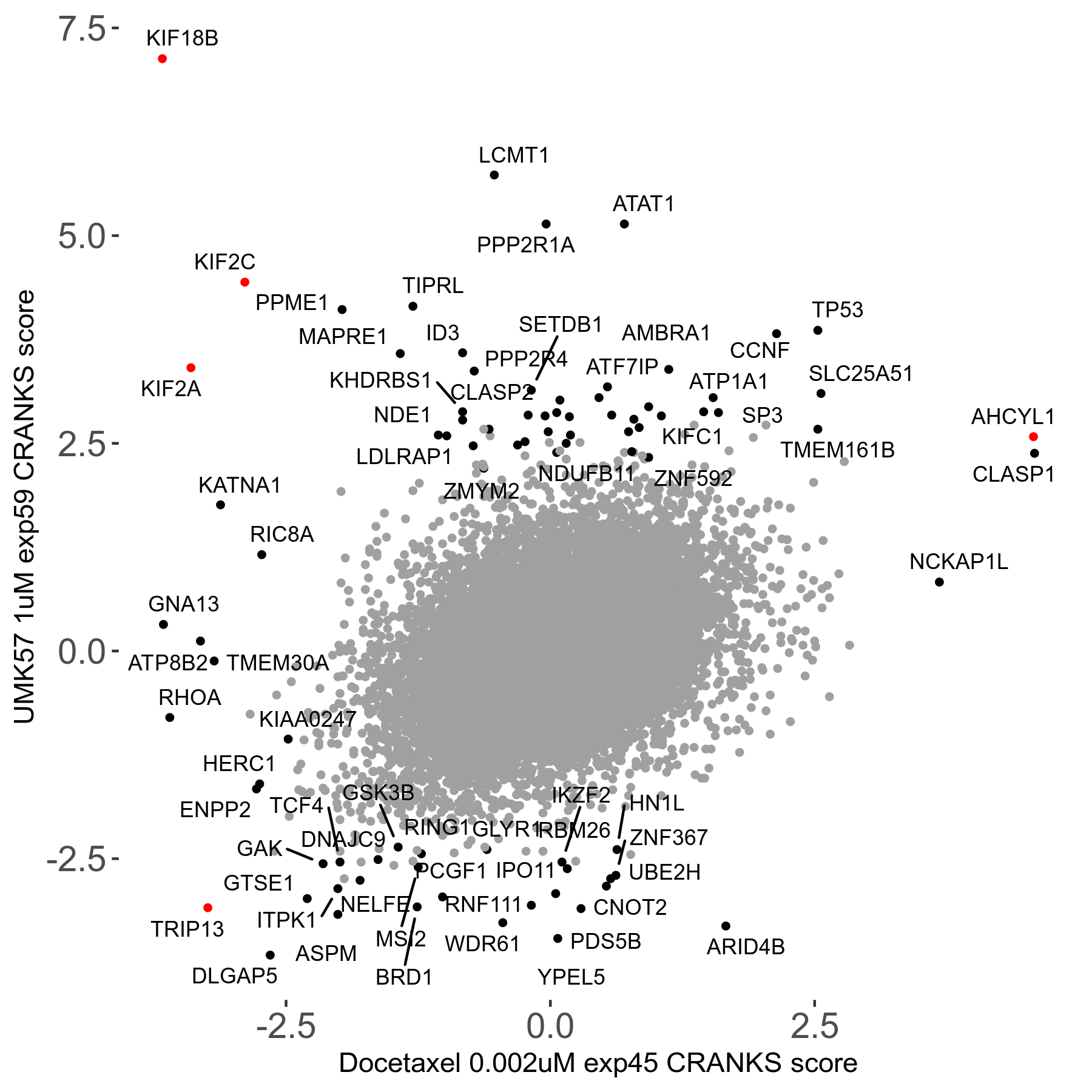
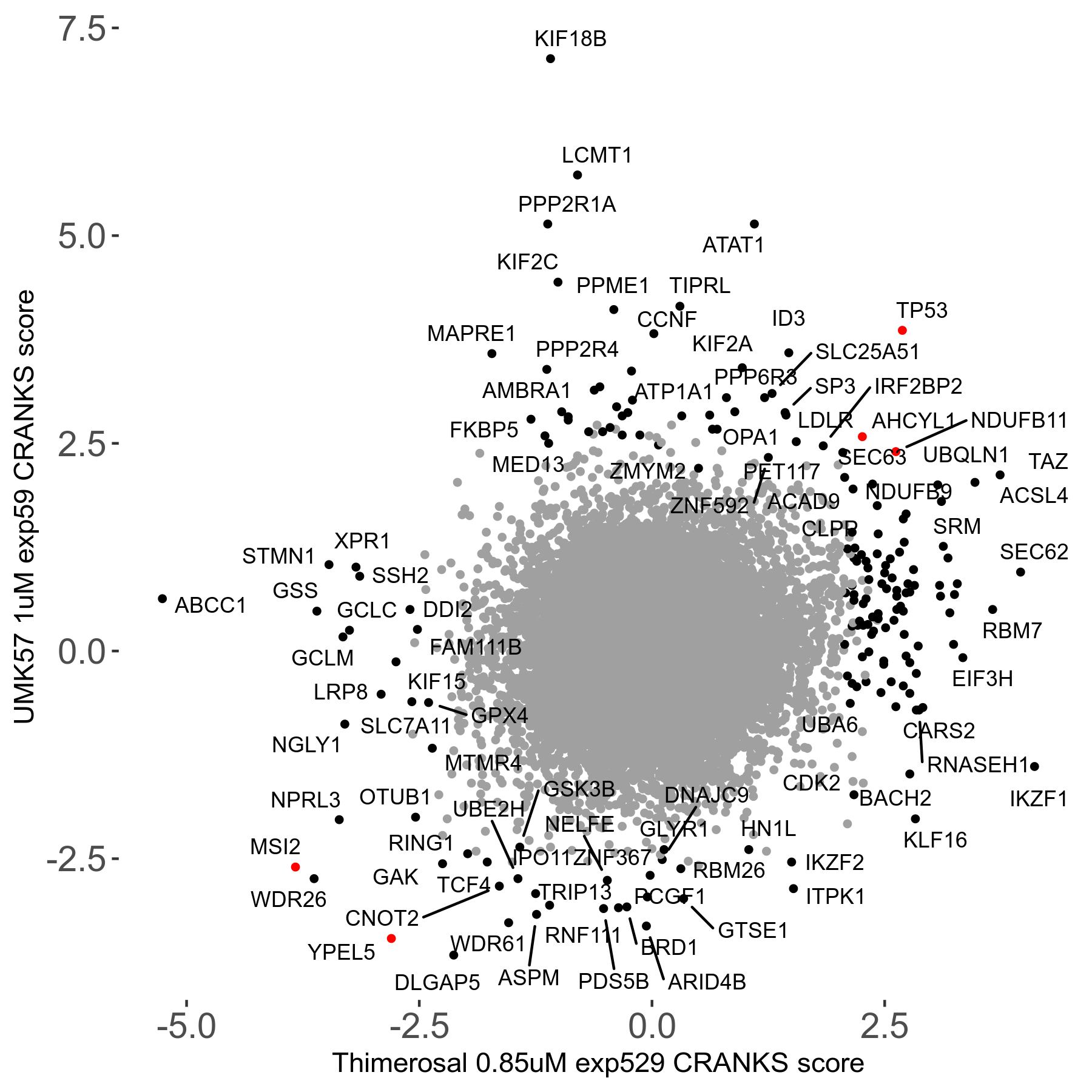
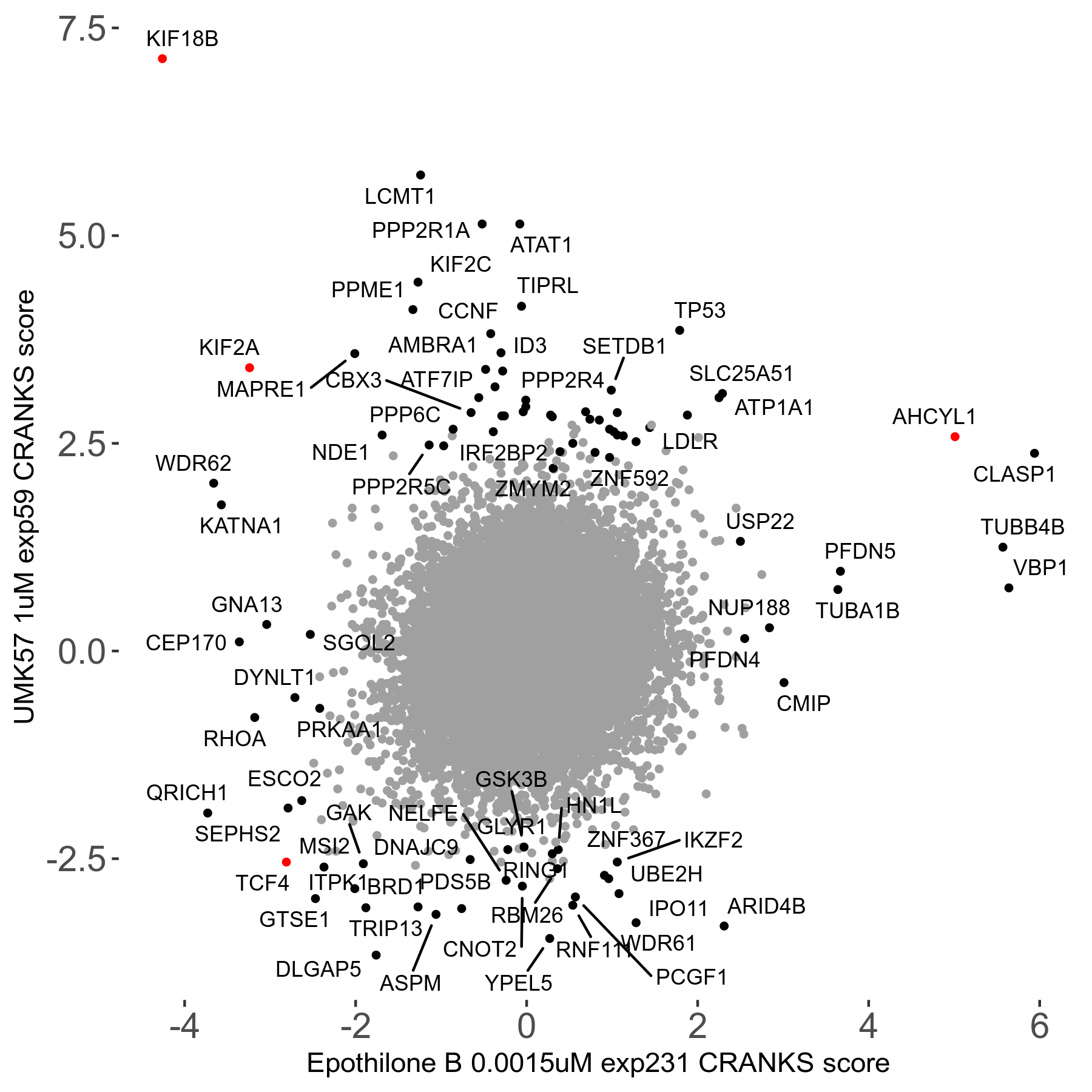
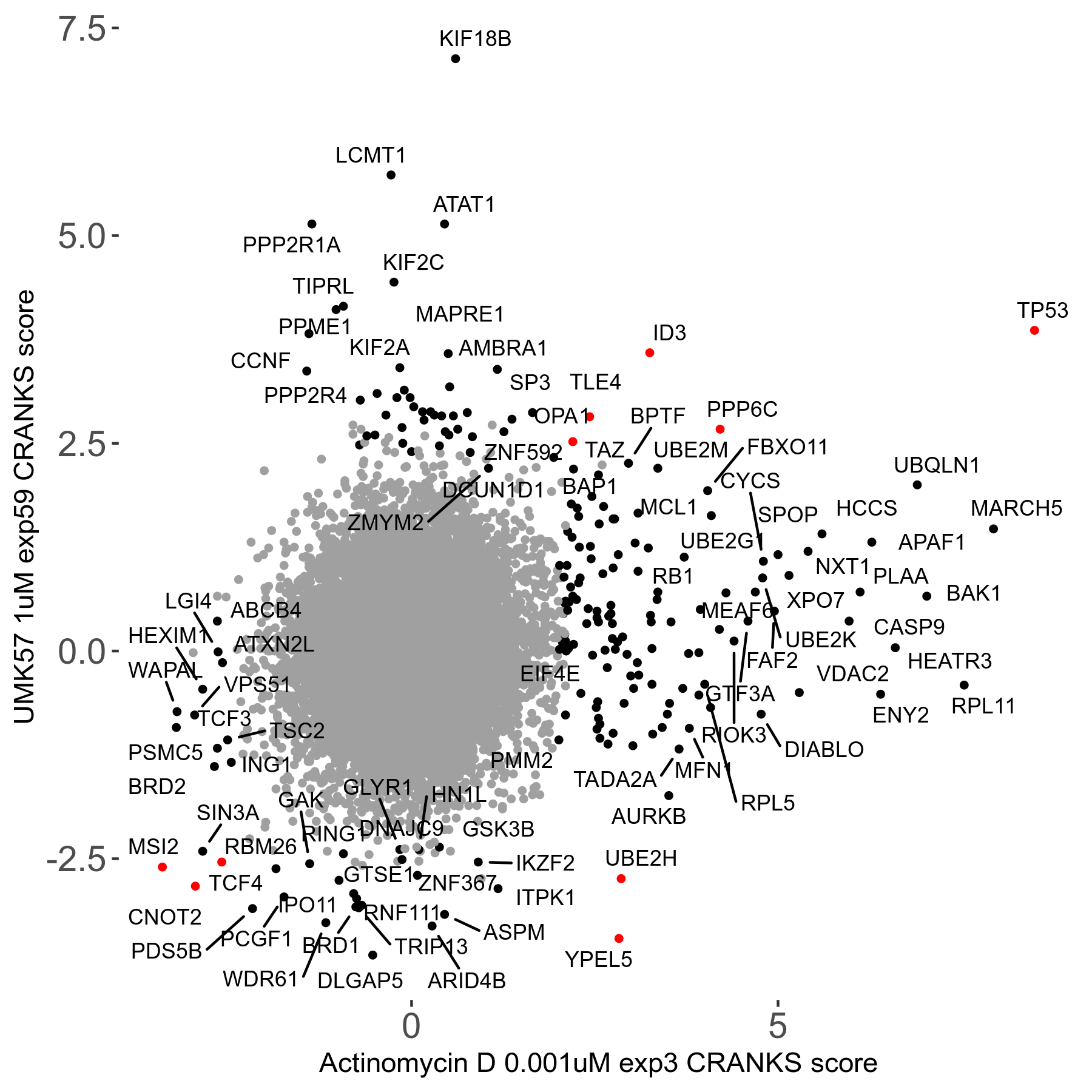
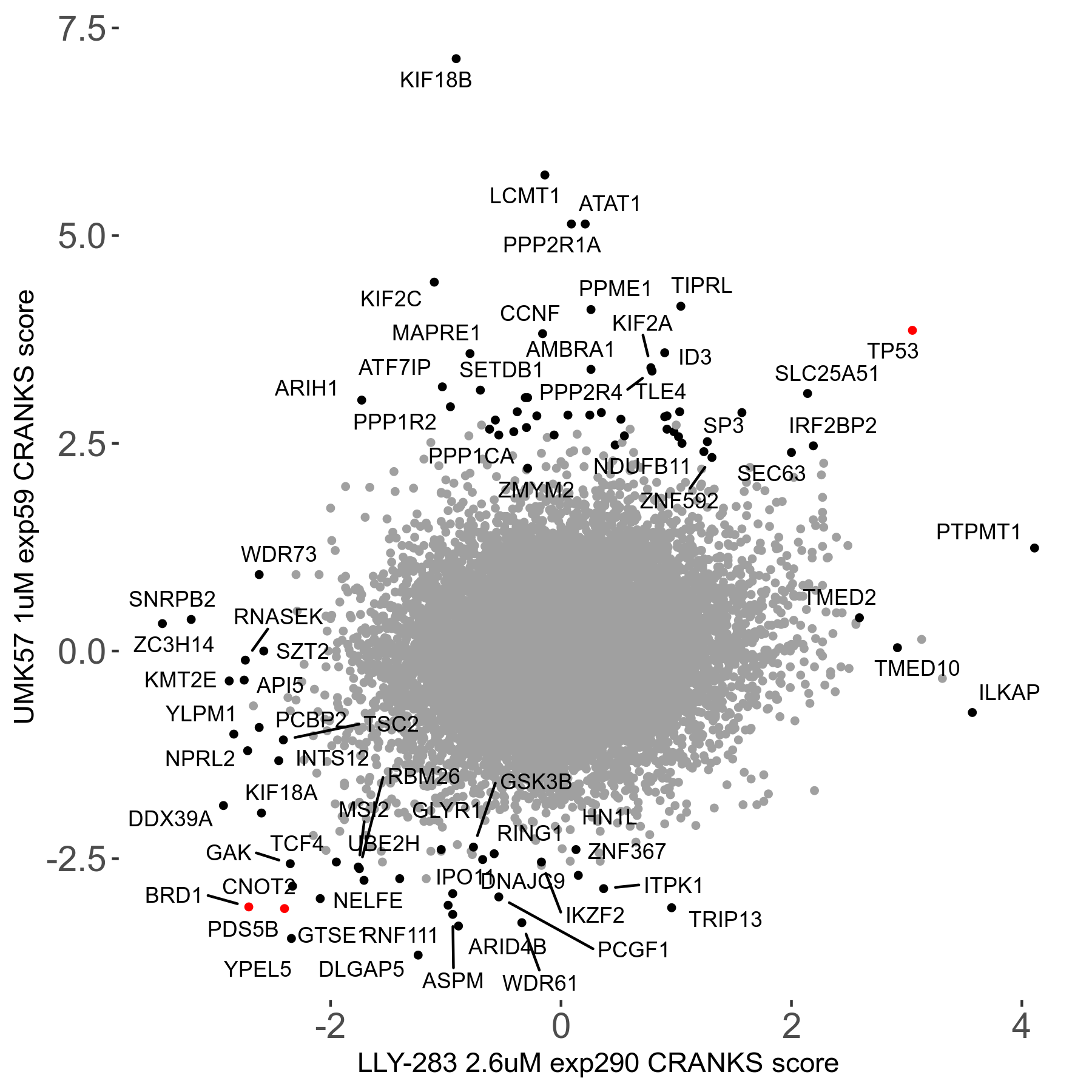
UMK57 1μM R01 exp59
Mechanism of Action
Potentiates kinesin-13 (MCAK), destabilizes kinetochore-MT attachment, suppresses chromosome mis-segregation
- Class / Subclass 1: Cell Cycle / Mitotic Inhibitor
- Class / Subclass 2: Organelle Function / Cytoskeletal Inhibitor
Technical Notes
Protein References
- PubChem Name: 5-(4-Methylphenyl)-4-pyrrolidinylthiopheno[2,3-d]pyrimidine
- Synonyms: N/A
- CAS #: 342595-74-8
- PubChem CID: 693939
- IUPAC: 5-(4-methylphenyl)-4-pyrrolidin-1-ylthieno[2,3-d]pyrimidine
- INCHI Name: InChI=1S/C17H17N3S/c1-12-4-6-13(7-5-12)14-10-21-17-15(14)16(18-11-19-17)20-8-2-3-9-20/h4-7,10-11H,2-3,8-9H2,1H3
- INCHI Key: VOAWQTDHSSKEKA-UHFFFAOYSA-N
- Molecular Weight: 295.4
- Canonical SMILES: CC1=CC=C(C=C1)C2=CSC3=NC=NC(=C23)N4CCCC4
- Isomeric SMILES: N/A
- Molecular Formula: C17H17N3S
Protein Supplier
- Supplier Name: In house - University of Montreal - Chemistry Platform
- Catalog #: N/A
- Lot #: N/A
Protein Characterization
- HRMS (ESI-TOF) m/z: [M+H]+ Calcd for C17H18N3S 296.1216; found 296.1142
Dose Response Curve
- Platform ID: UMK57
- Min: -2.8530; Max: 75.4945

| IC | Concentration (µM) |
|---|---|
| IC10 | N/A |
| IC20 | 0.3698 |
| IC30 | 0.6436 |
| IC40 | 1.0136 |
| IC50 | 1.5379 |
| IC60 | N/A |
| IC70 | N/A |
| IC80 | N/A |
| IC90 | N/A |
Screen Summary
- Round: 01
- Dose: 1µM
- Days of incubation: 8
- Doublings: 5.2
- Numbers of reads: 15049192
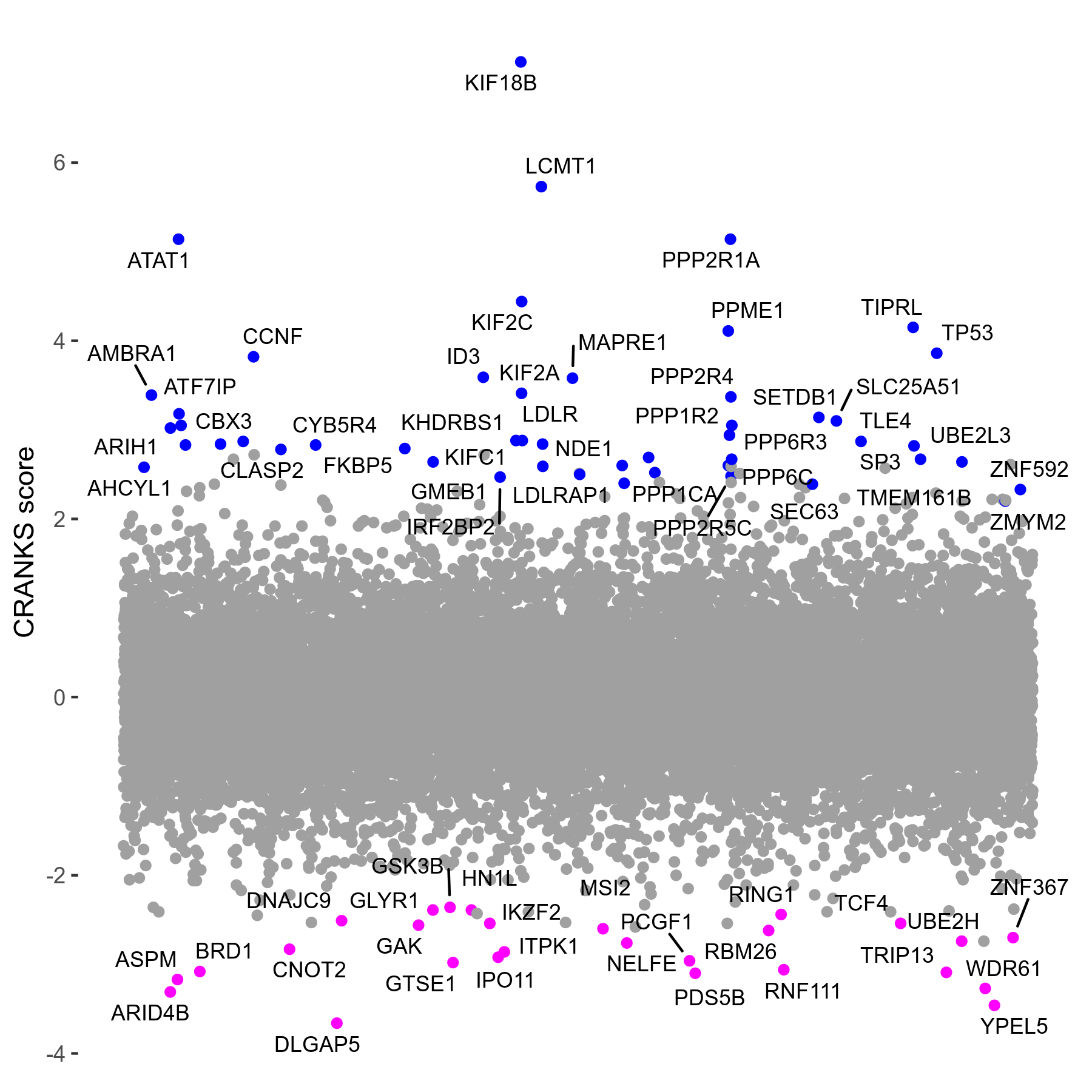
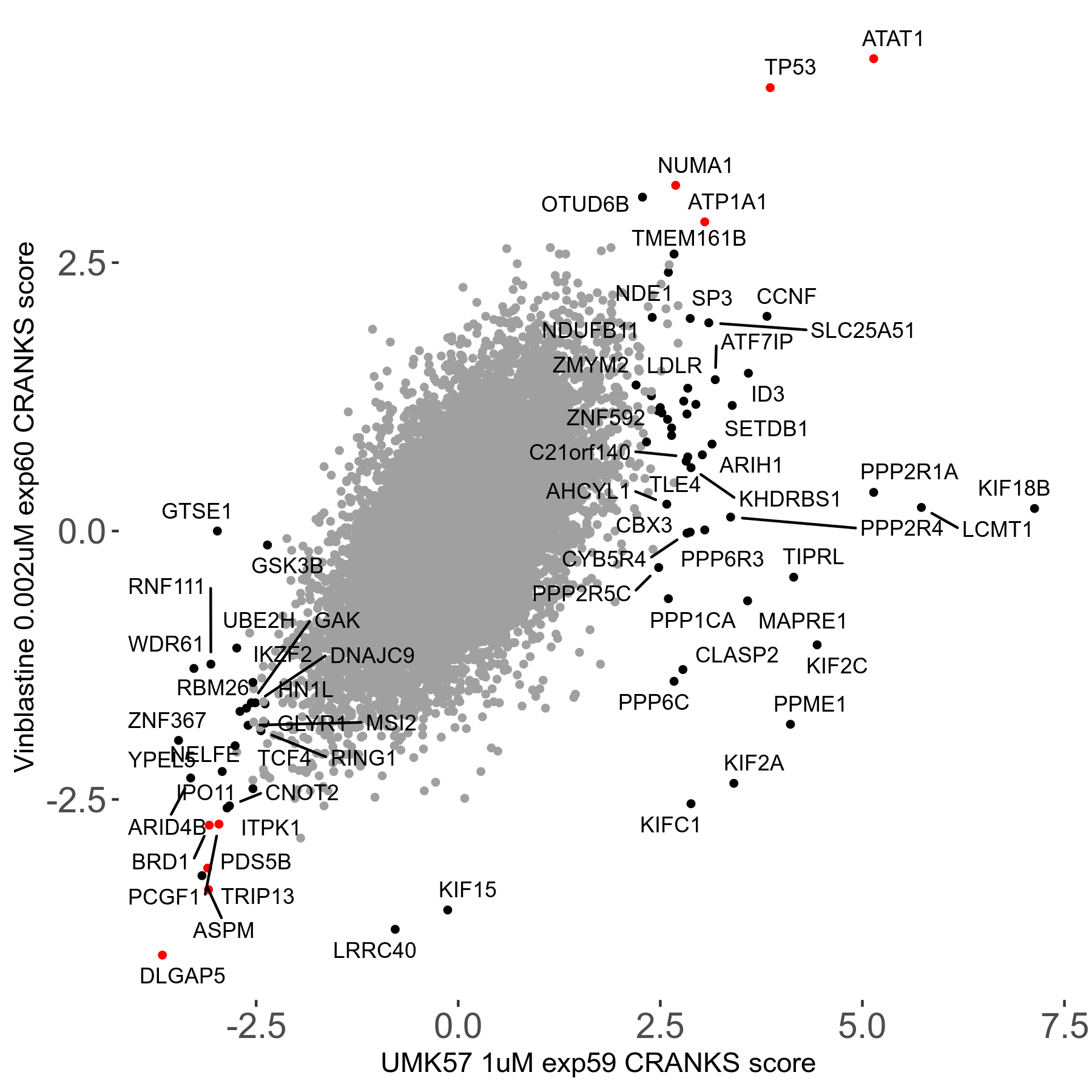
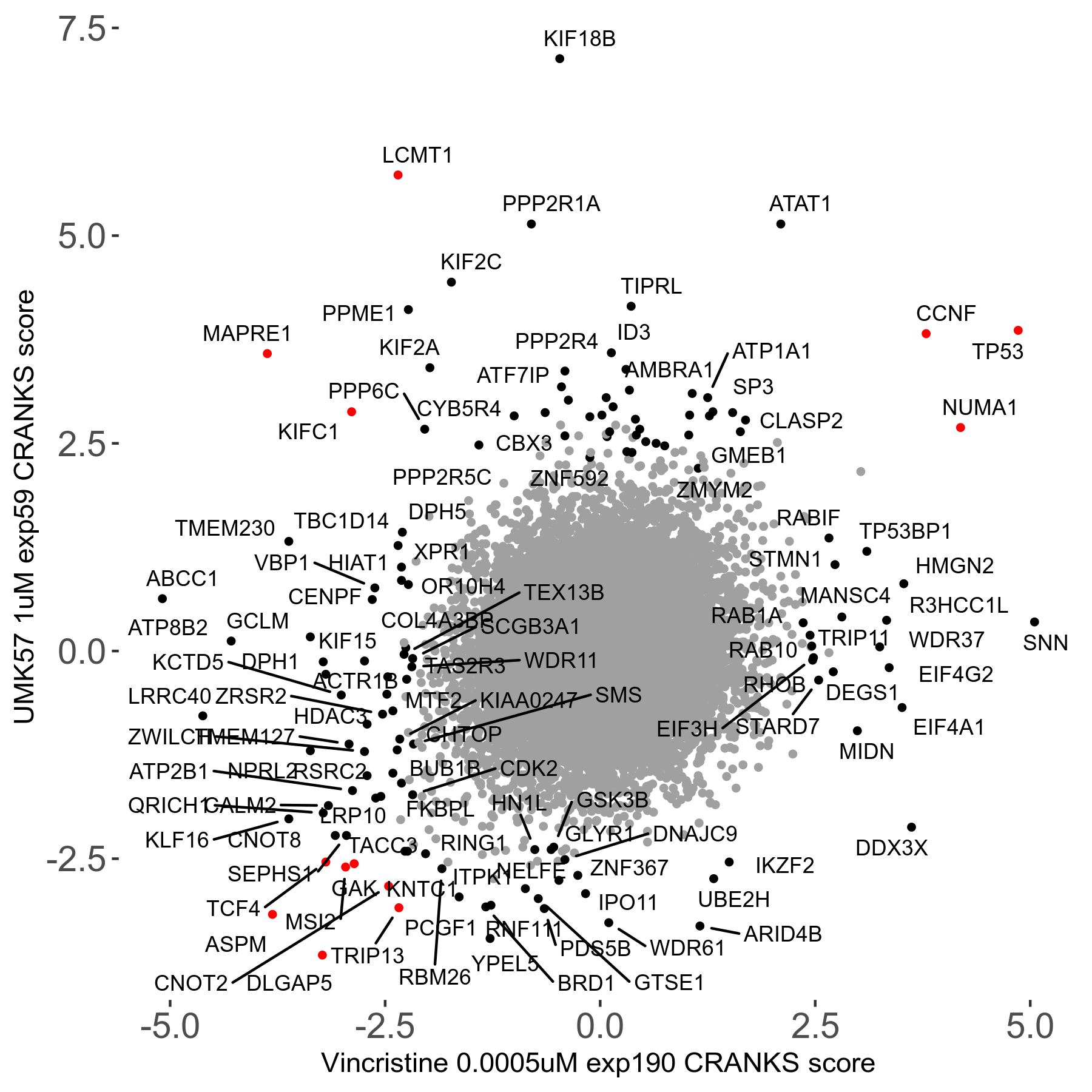
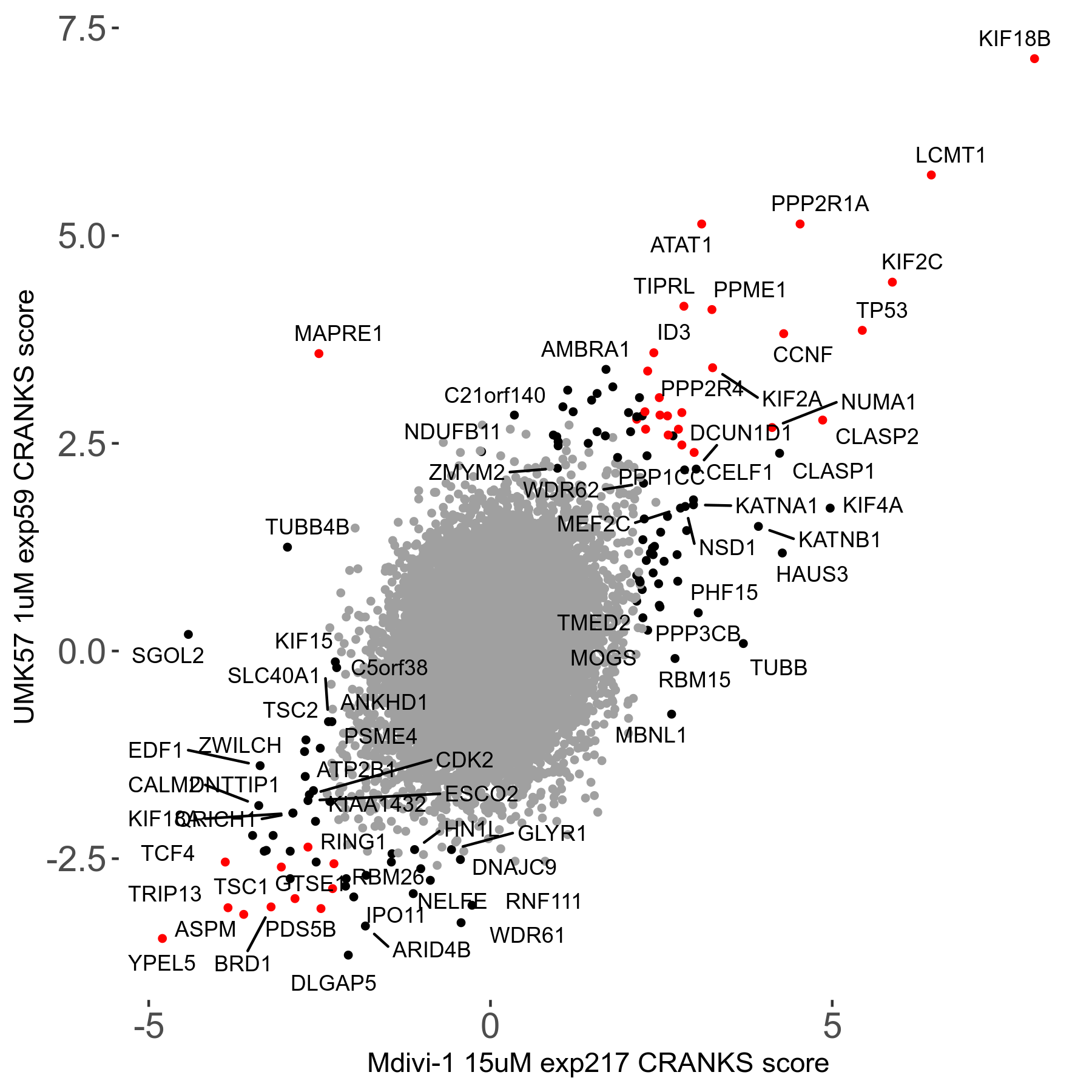
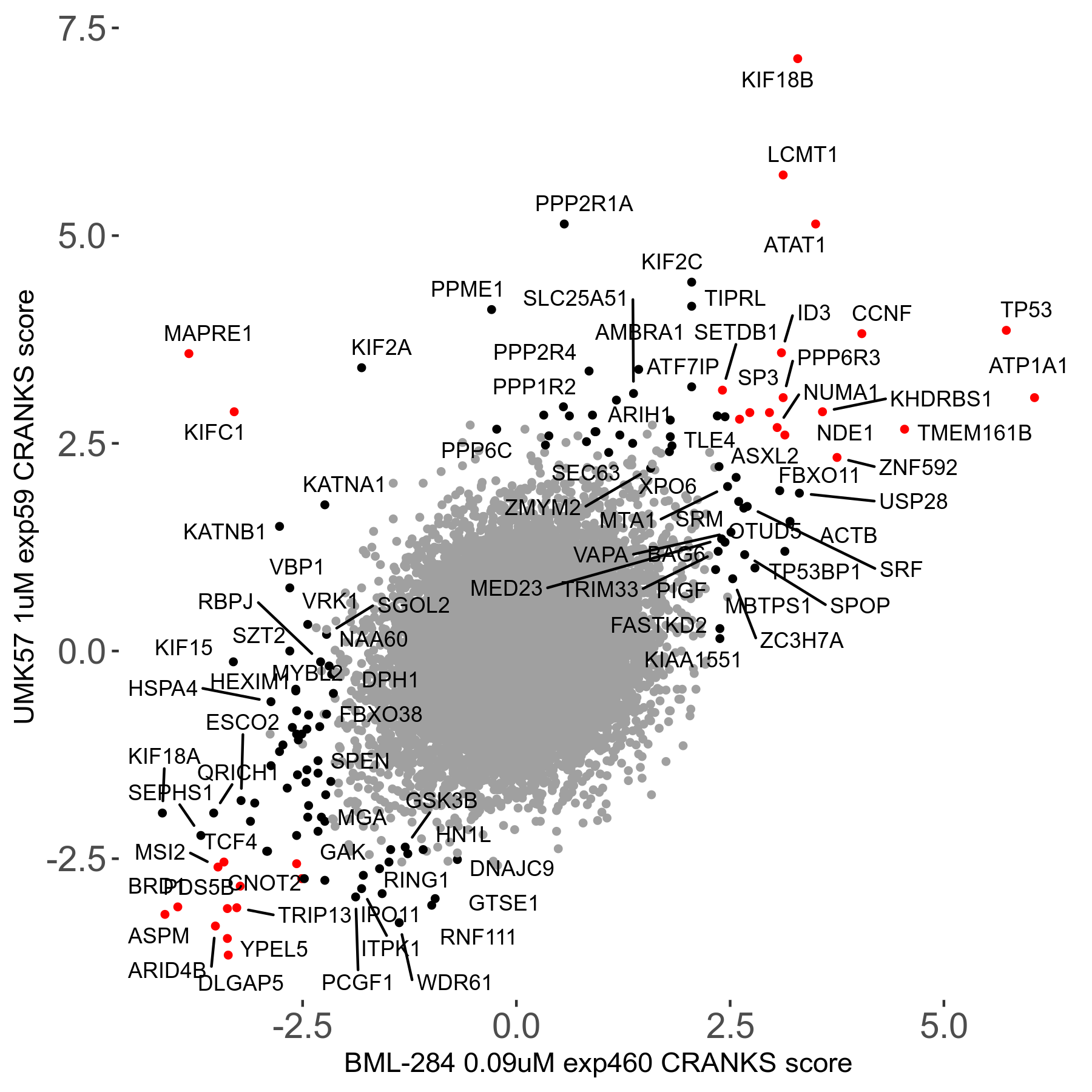
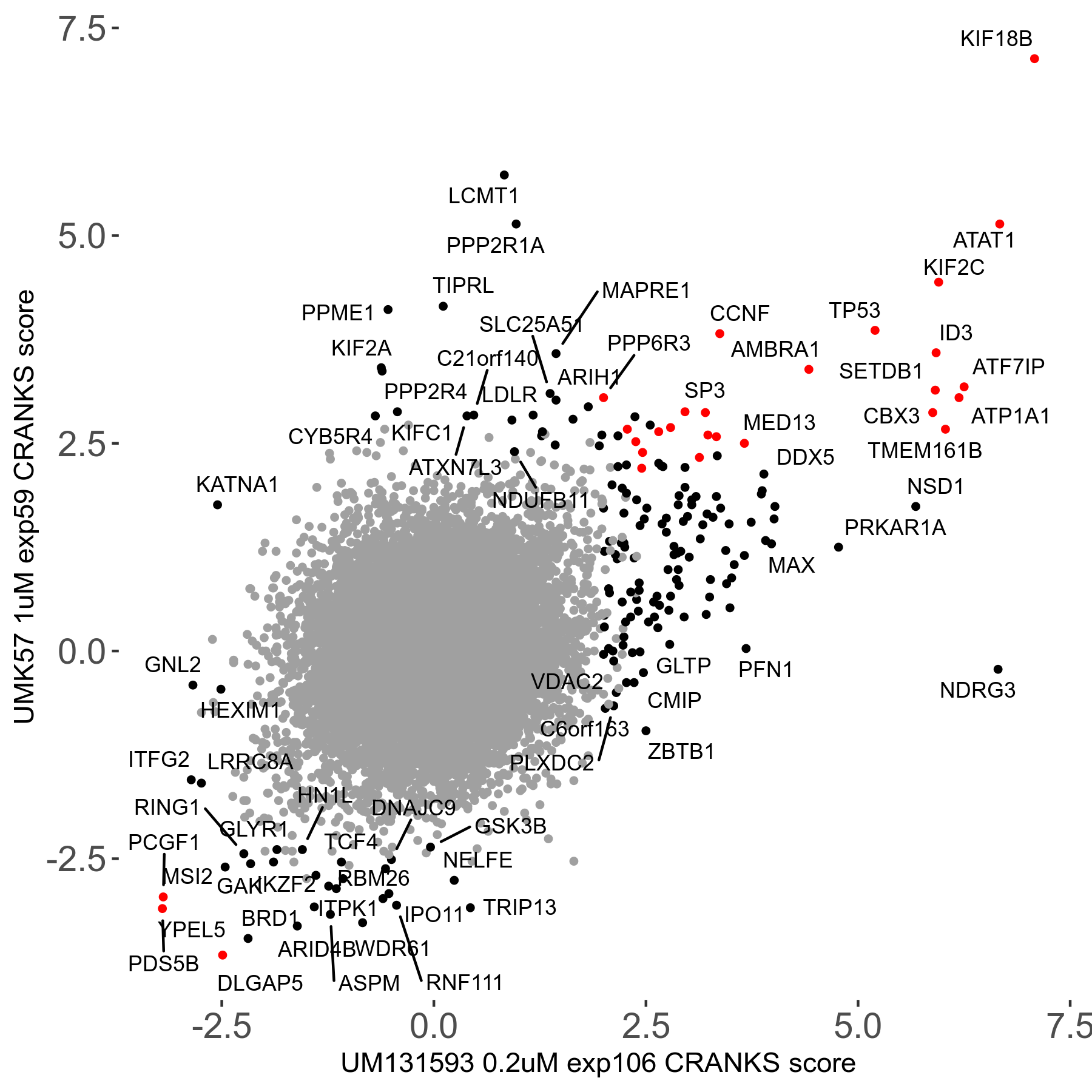
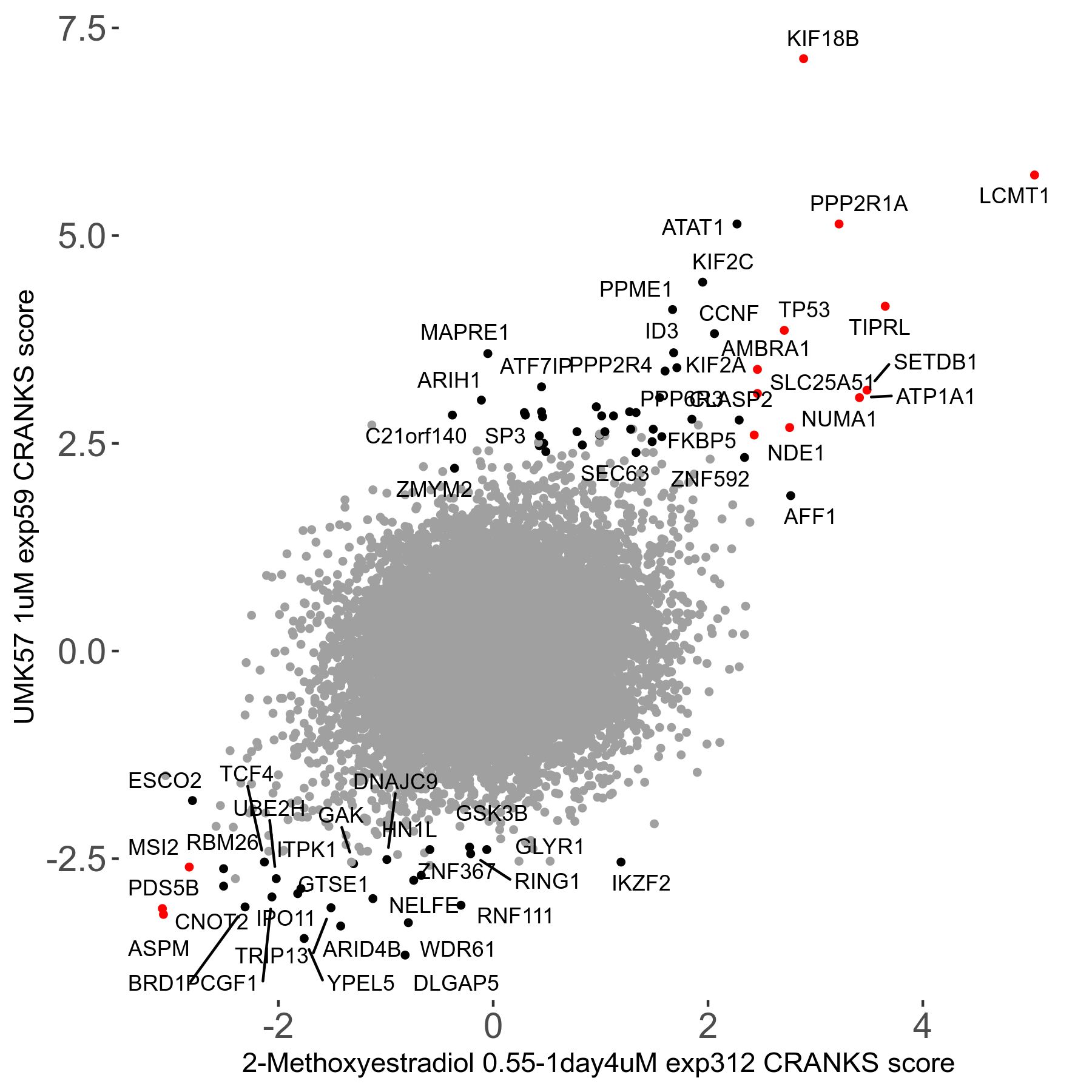
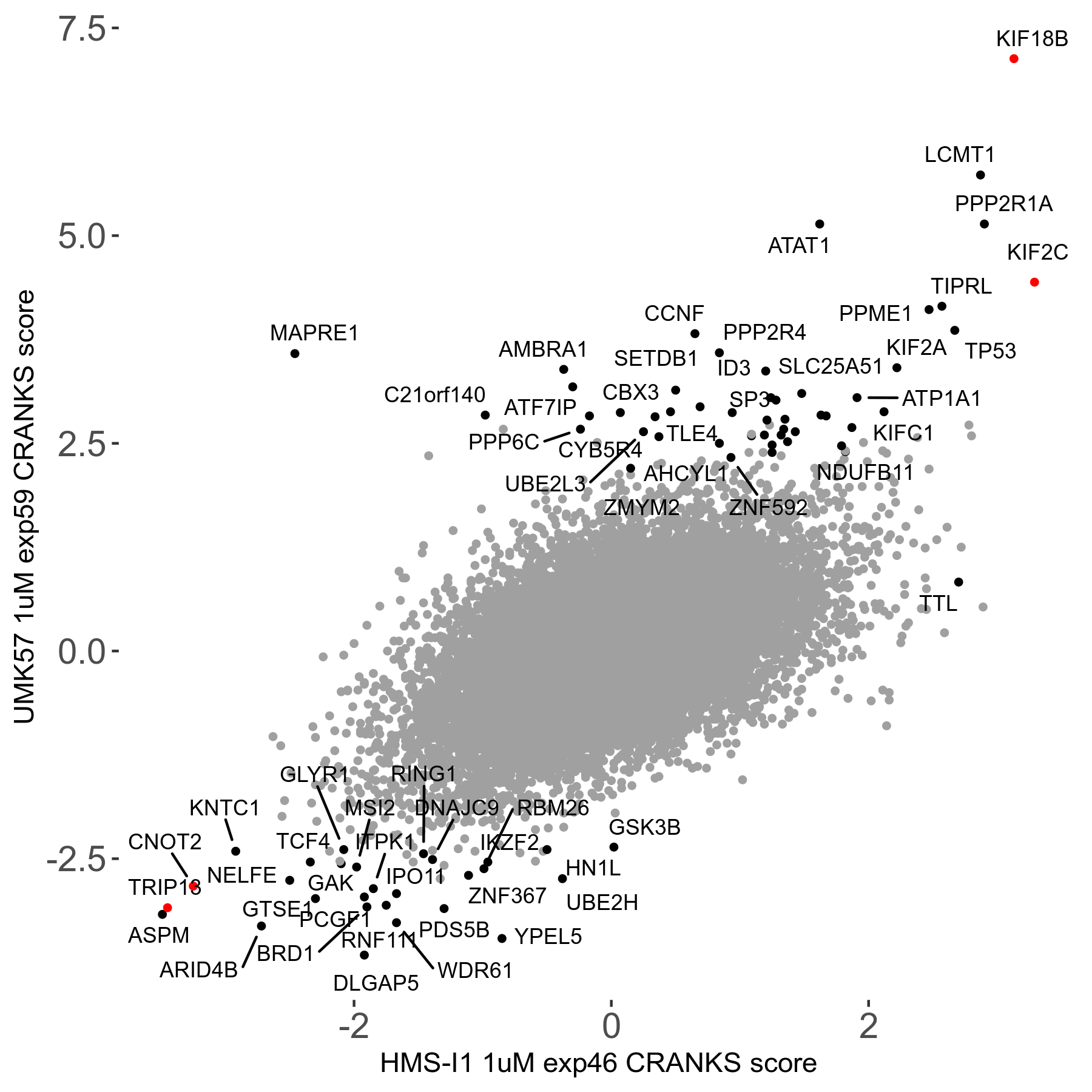
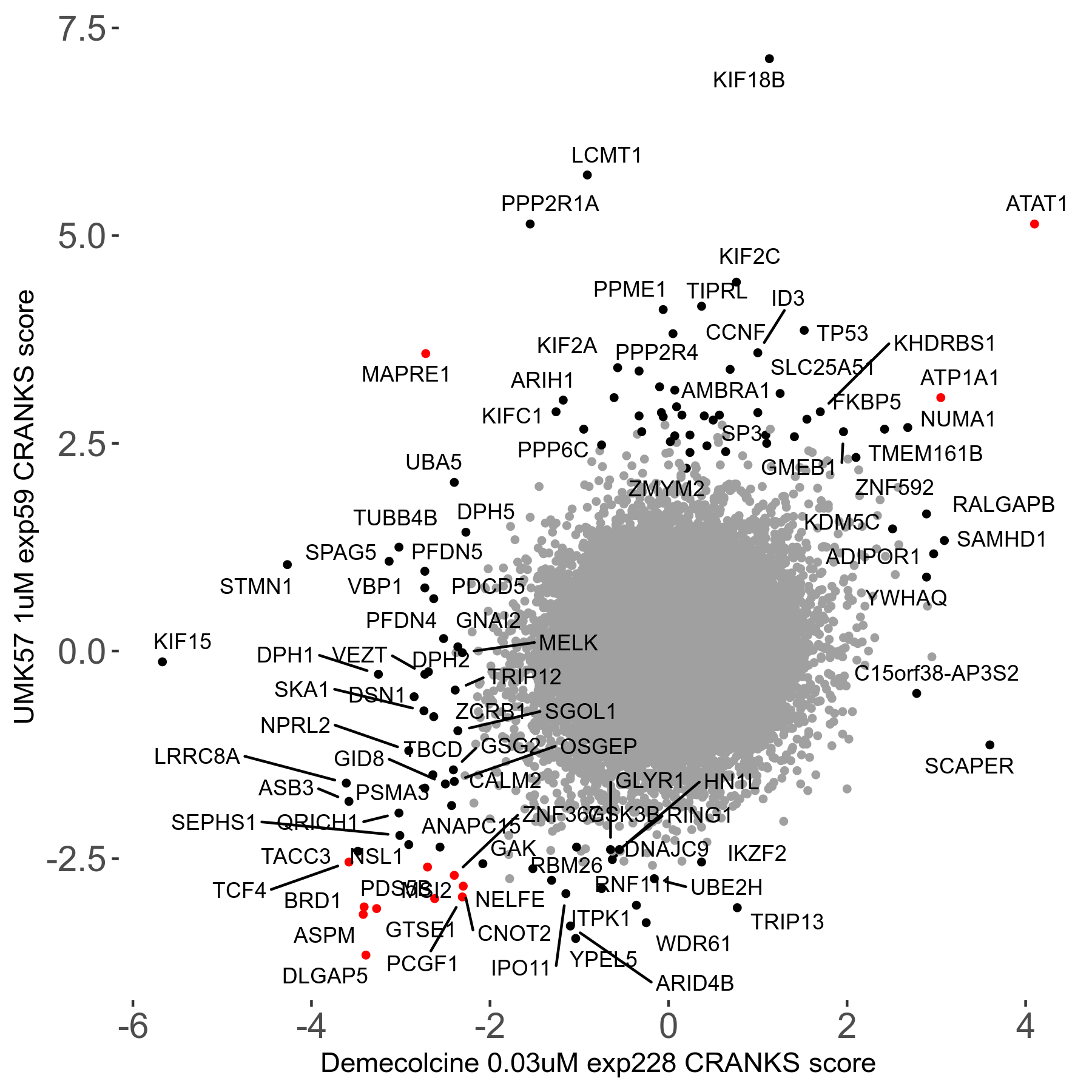
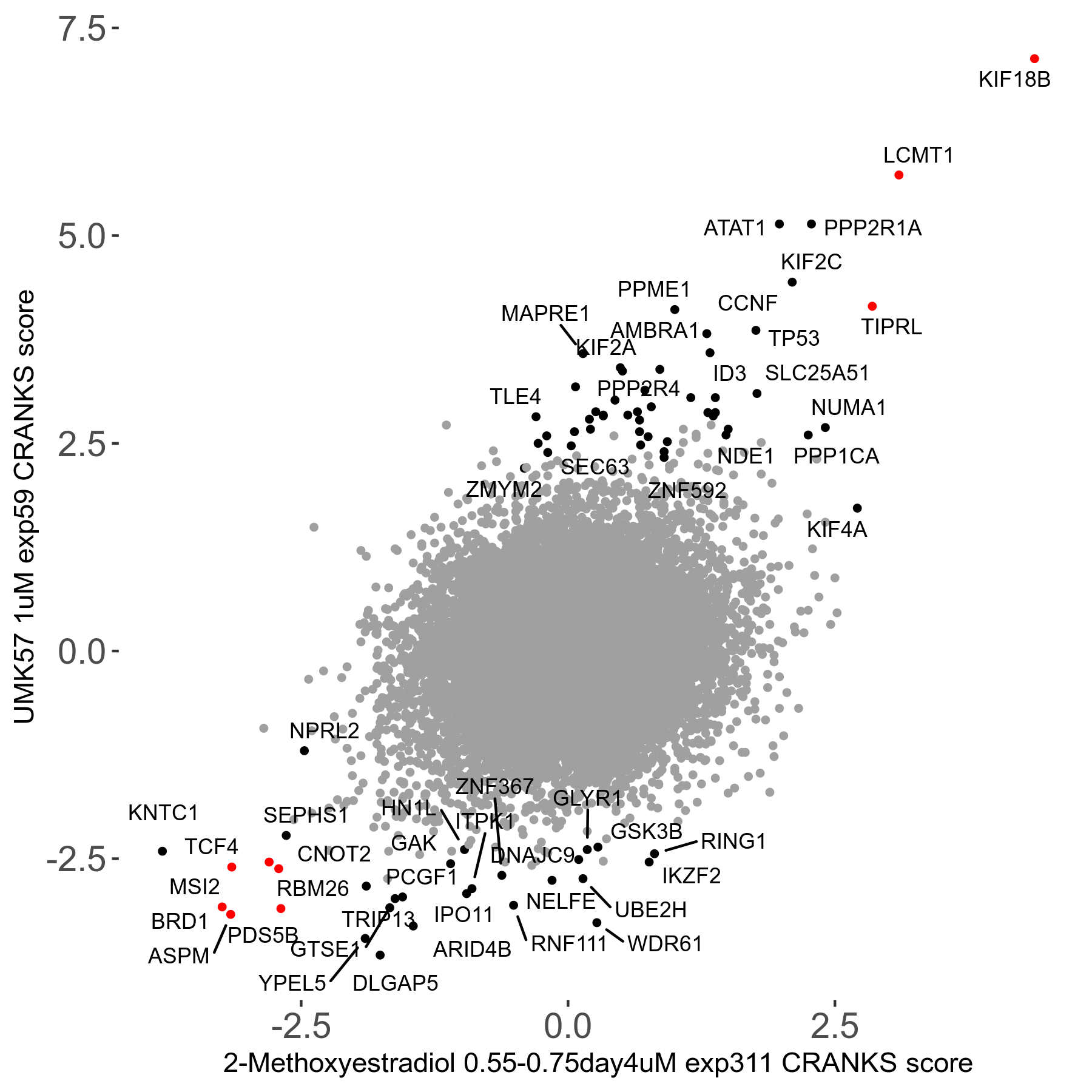
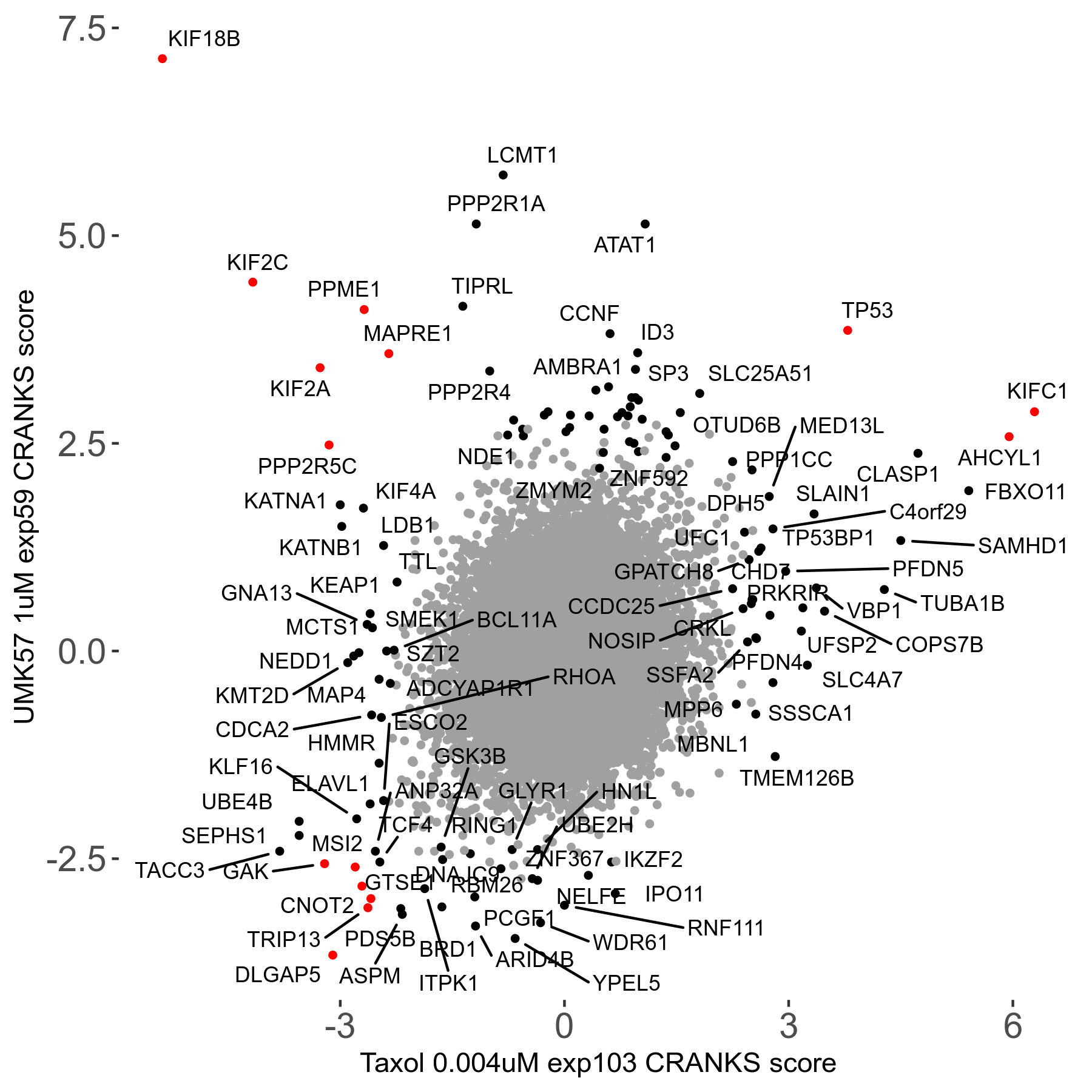
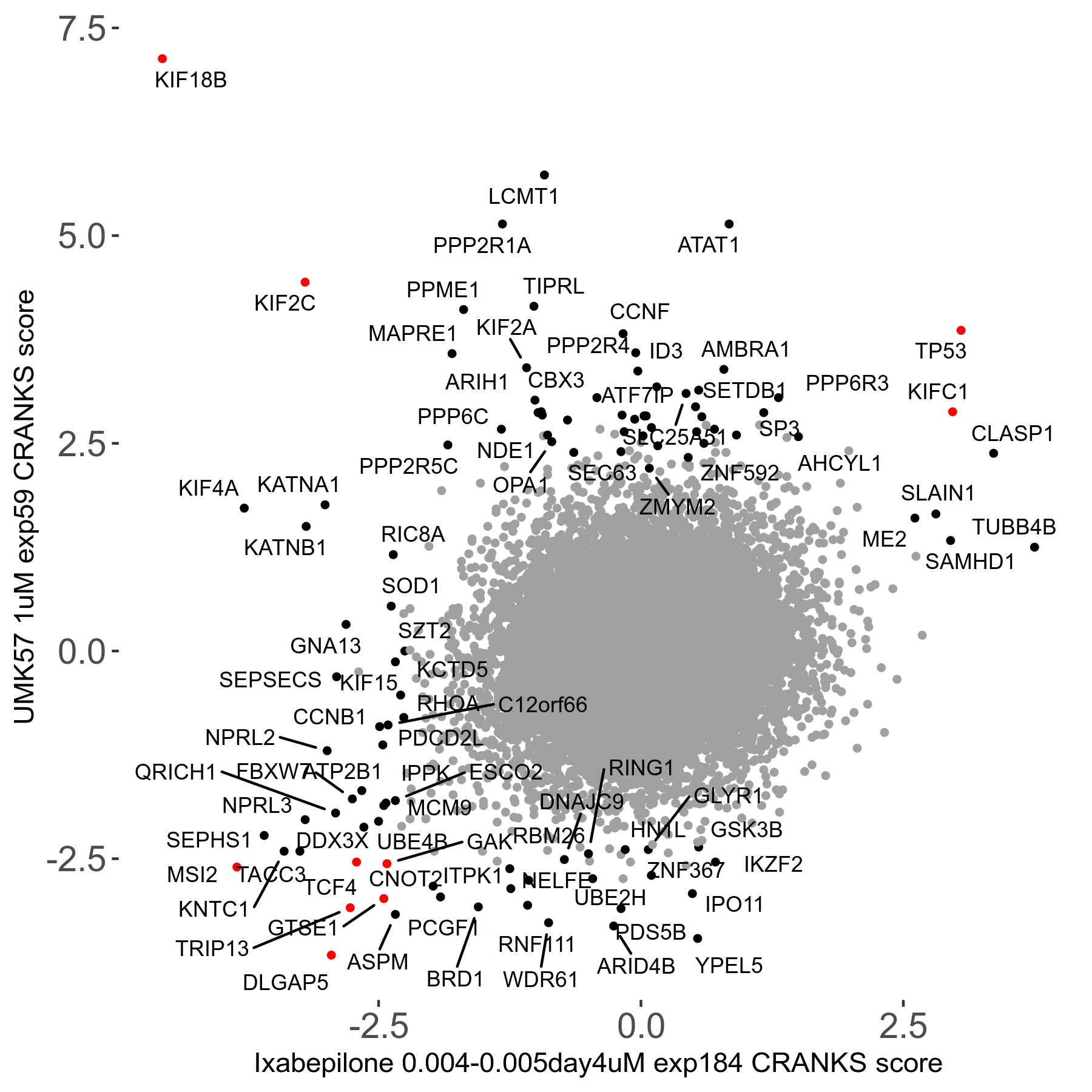
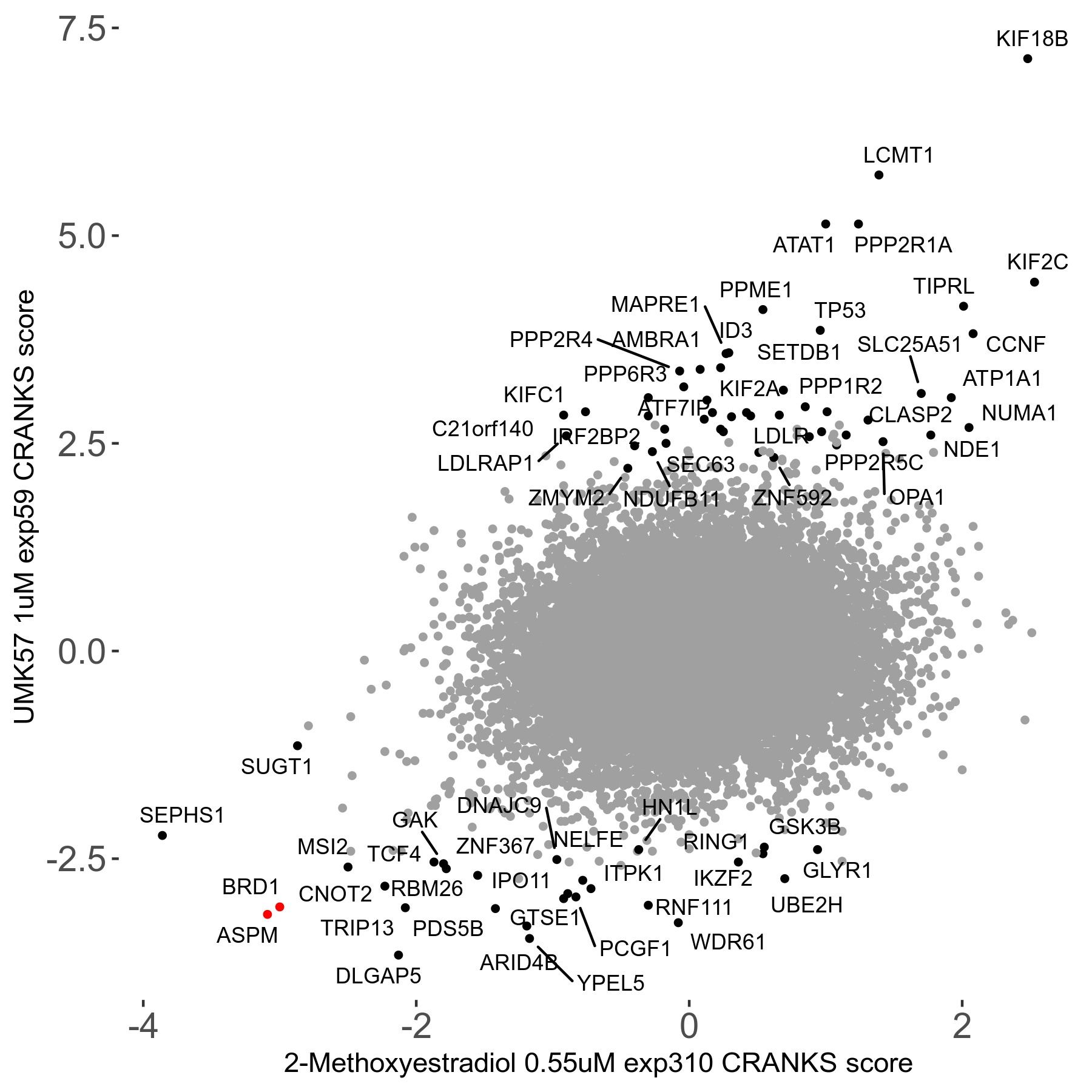
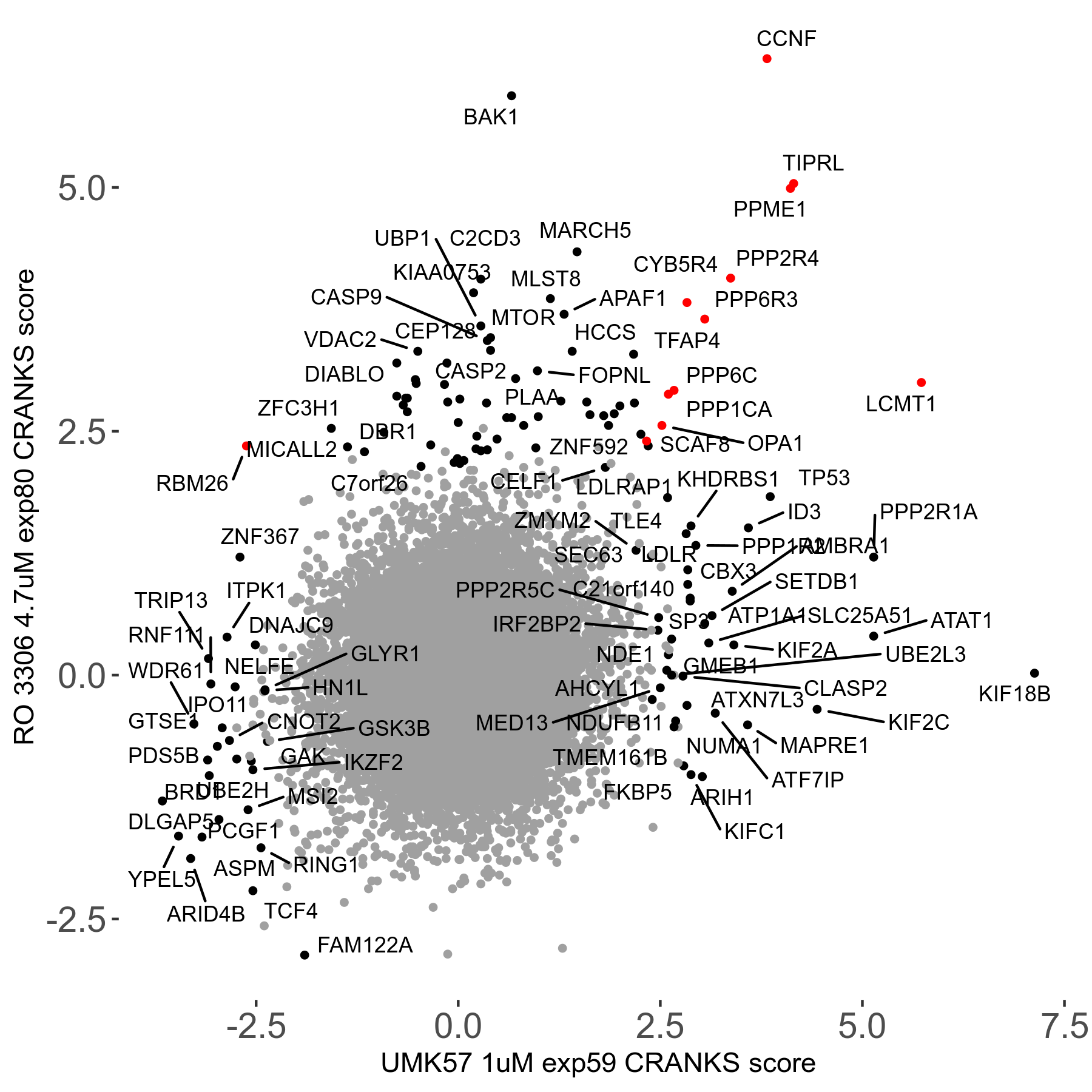
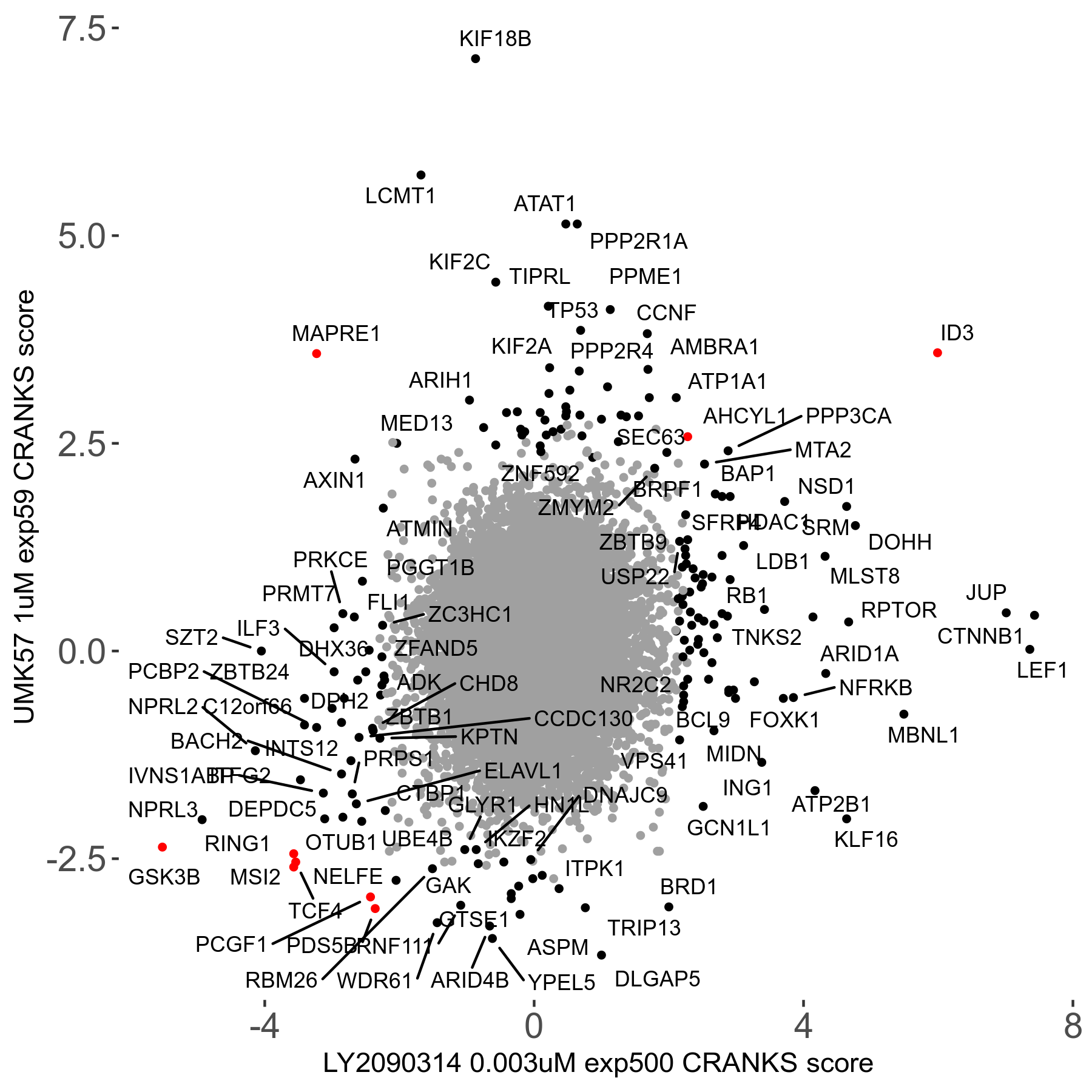
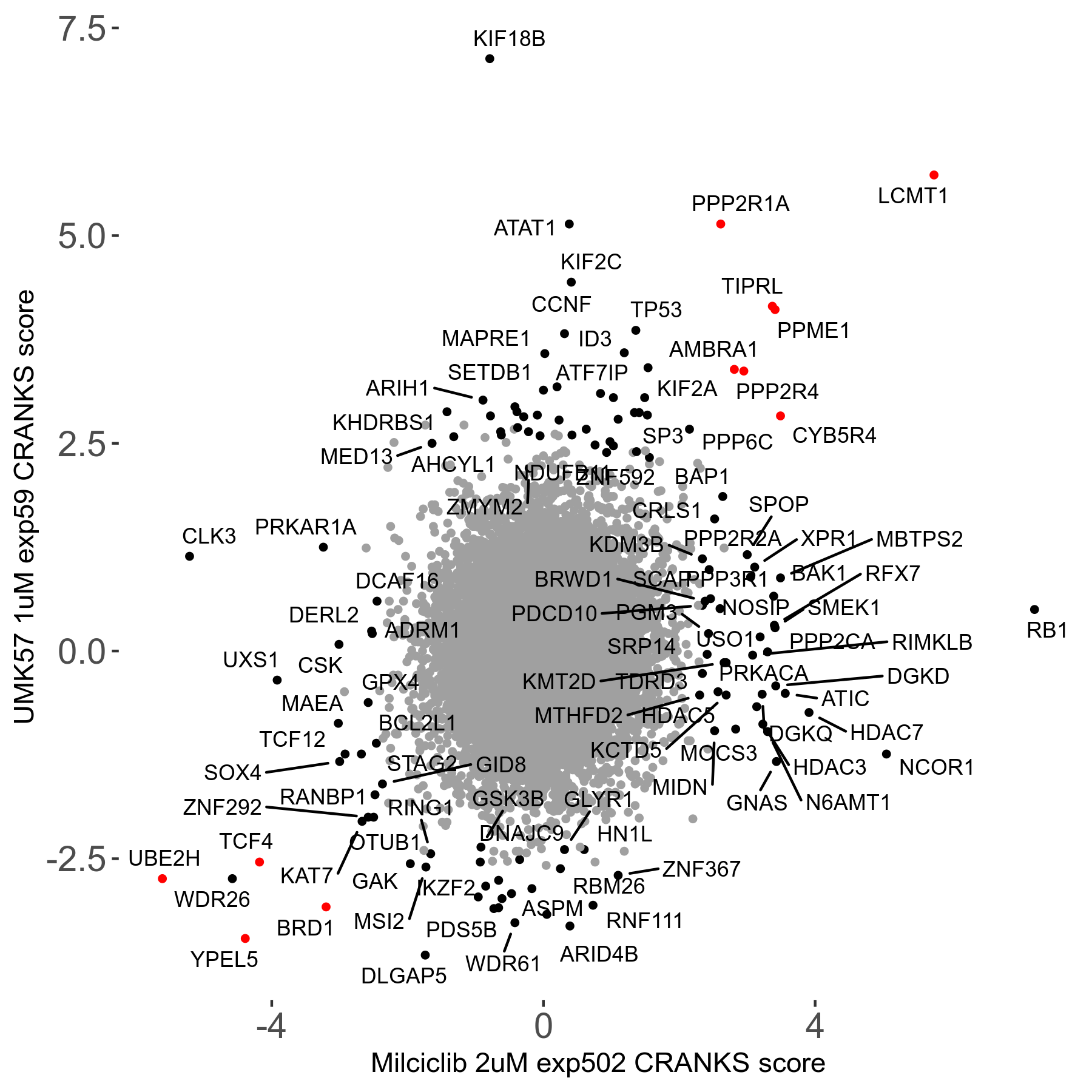
Screen Results
| Sensitive/Resistant hits (FDR<0.05) | CRANKS | Score Plot | Top 30 Genes | Screen Similarity | Top 30 Sensitive GO terms | Top 30 Resistant GO terms |
|---|---|---|---|---|---|---|
| 27/49 | Scores |